Course preparation: R basics
Here, we aim to provide a brief overview on the basics of R, which is supposed to refresh your knowledge and help you to identify topics worth revisiting. You are encouraged to refresh your knowledge from the module “Quantitative Methods for Geographers” as well as learn from online resources we list at the bottom of this page.
Operators
R is an interpreted programming language which means users can execute commands directly in the console without having to compile sourcecode. You can use R as a simple calculator using arithmetic operators and mathematical functions which are then evaluated directly. For example:
# A simple calculation
2*(1+4)
# abs() returns absolute values
abs(-2)
# sqrt() returns the squareroot
sqrt(pi) # Note: 'pi' is a predefined constant## [1] 10
## [1] 2
## [1] 1.772454To list the defined operators used by R, simply type
?Syntax into the console. This includes the arithmetic
operators
- addition
+and subtraction- - multiplication
*and division/ - exponentiation
^or**
as well as logical operators
- less than
</ greater than> - less than or equal to
<=/ greater than equal to>= - equal to
== - not equal to
!= - or
| - and
&
The concept of logical operators is to allow a program to make a
decision based on (multiple) conditions. A condition can be evaluated to
be either TRUE or FALSE (see
?Logic). For example:
2 > 5## [1] FALSEThey are often used in combination with control statements such as
the conditional if...else statement which are mentioned
below. But first back to basic functions: R also comes with a whole set
of mathematical functions including
- trigonometric functions
sin(),cos()andtan() - exponential function
exp() - statistical metrics like
min(),max(),median()
to name just a very few of them. Functions in R provide information
on how to use them. If you need help with a function, type in
help(name_of_function) or simply
?name_of_function to display the correct usage and syntax
of a function.
Objects
Everything in R we assign to a variable is an object. We can assign
objects in R with the <- operator (i.e. an arrow),
e.g. x <- 2. Commonly, other languages use
= which also works in R, however it is definitely good
practice to use the <- operator in R.
# Assign object 'x' a value of 2
x <- 2
# <- assigns the value, but does not print the result to console
# Assign another variable 'y' ...
y <- 5
# Store the product of x and y in a new object 'z'
z <- x*y
# Print z
print(z)## [1] 10Assigned variables are stored in the environment or
workspace of our session and can be listed by calling the
ls() function.
Data Types and Data Structures
The basic data types in R comprise:
- Character (or String):
sensor <- 'Landsat-8' - Integer:
n_bands <- 11L(theLtells R it is an integer, otherwise it is interpreted as numeric) - Numeric (or Float):
lambda_red <- 0.662 - Logical (or Boolean):
sun_synchronous <- TRUEorsun_synchronous <- T - Factors:
agency <- factor('NASA',levels=c('NASA','ESA','INPE'))
Understanding data types is important for a variety of reasons. For
instance, later in the course we will manipulate large rasters of
remotely sensed reflectance data which may be stored as floating point
numbers between 0 and 1. However, storing
raster matrices as floats on disk requires more memory as compared to
using an integer format. It can therefore be handy to transform data
into integer prior to saving it to disk.
For now, let us just keep in mind that there are different data types and if we now want to store not just a single value into a variable but a whole set of data, we need to undertstand the different data structures available in R:
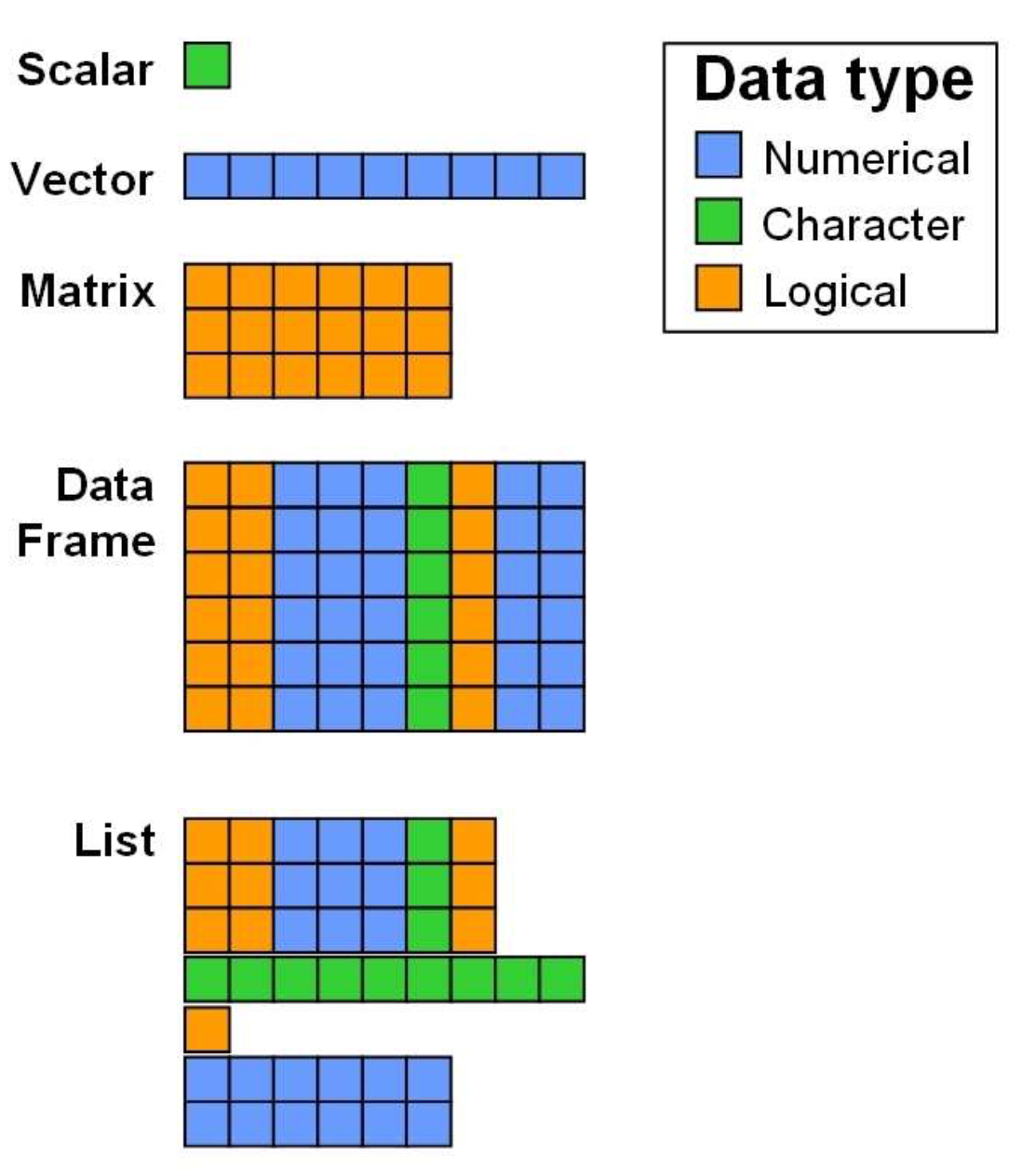
Data Structures in R (from Zimmermann & Steinmann 2006)
As you can see in the figure above, a variety of data structures
exist in R, which either allow for storing different data types
simultaneously (data frames, lists), or only
one datatype (scalars, vectors,
matrices). Note, that the colors representing the data
types are only chosen for vizualisation purposes: It does not mean that
matrices or vectors can only store values of type logical or numeric,
but only data of the same type.
Vectors
A vector in R can be created using a variety of functions. The most
basic function is the c()-function, where ‘c’ stands for
concatenate.
# Create a vector of numbers
a <- c(2, 4, 8, 16, 32)
# Print vector a in console
print(a)
# cat-function: Similar to print, outputs the objects, concatenating the representations
cat('vector a:', a) ## [1] 2 4 8 16 32
## vector a: 2 4 8 16 32Let`s see what happens when we mix numeric values with strings and combine them into a vector.
b <- c(2, 4, 8, 16, 32, 'Landsat', 'Sentinel-2')
print(typeof(b))## [1] "character"Our vector was automatically cast to character. This is
because R will try and convert a smaller data type into a larger one to
avoid data loss (here from numeric to
character). The strings (Landsat and
Sentinel-2) in vector b cannot be cast to
numeric, however we can very well transform a numeric 2
into a string '2'. Be aware of this behaviour and use other
data structures if needed.
Back to vector creation. Instead of having to type in sequences or
replicates manually by hand, R provides the functions seq()
and rep().
# Create a numeric sequence and store in object s
s <- seq(from=1, to=365, by=1) # or simply s <- seq(1, 365, 1)
cat('Length of s:', length(s))
# Alternatively use (always with by=1):
s <- c(1:365)
# We may want to repeat a value n times...
r <- rep(1, 5)
print(r)
# ...or repeat a sequence
r <- rep(seq(2, 8, 2), 3)
print(r)
# ...or repeat each entry in a sequence
r <- rep(seq(2, 8, 2), each=3)
print(r)## Length of s: 365[1] 1 1 1 1 1
## [1] 2 4 6 8 2 4 6 8 2 4 6 8
## [1] 2 2 2 4 4 4 6 6 6 8 8 8Matrices and Arrays
Creating matrices and arrays is just as easy as creating vectors.
Matrices are just 2-dimensional arrays with a specified number of
rows and columns. All columns in a matrix must
have the same data type (e.g., numeric, character) and must be of equal
length.
# Creates empty matrix of size 5x2
m1 <- matrix(data=NA, nrow=5, ncol=2)
print(m1)
# Creates matrix of size 5x2 containing a numeric vector
m2<- matrix(data=c(1:10), nrow=5, ncol=2)
print(m2)
# Can you describe what byrow=T does?
m3 <- matrix(data=c(1:10), nrow=5, ncol=2, byrow = T)
print(m3)## [,1] [,2]
## [1,] NA NA
## [2,] NA NA
## [3,] NA NA
## [4,] NA NA
## [5,] NA NA
## [,1] [,2]
## [1,] 1 6
## [2,] 2 7
## [3,] 3 8
## [4,] 4 9
## [5,] 5 10
## [,1] [,2]
## [1,] 1 2
## [2,] 3 4
## [3,] 5 6
## [4,] 7 8
## [5,] 9 10Arrays are similar to matrices, but can have more than two
dimensions, as specified in the dim argument.
# Create array of two 4x3 matrices
a <- array(data=c(1:12), dim=c(4,3,2))
print(a)## , , 1
##
## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 10
## [3,] 3 7 11
## [4,] 4 8 12
##
## , , 2
##
## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 10
## [3,] 3 7 11
## [4,] 4 8 12In R, basic mathematical operations may be used to manipulate arrays. This can be used to add, subtract, multiply or divide a constant from every element in the array.
# Create array of one 4x3 matrices
a <- array(data=c(1:9), dim=c(4,3,1))
a2 <- a * 10An entire array can be scaled by multiplying it with a constant
We can also use these operations on multiple arrays of the same size. This array arithmetic works element-wise. For example multiplying two arrays together, the first element element of the first array is multiplied by the first element of the second array, and so on. Note: while not needed in this course, R is also capable of performing matrix algebra using these operators.
# Create two arrays each of one 4x3 matrices
a1 <- array(data=c(1:9), dim=c(4,3,1))
a2 <- array(data=c(1:9), dim=c(4,3,1))
a3 <- a1 * a2Multiplying two arrays of the same shape together results in element-wise multiplication
Matrices are very important structures when working with remote
sensing data, as this is how images are represented numerically. We will
extensively make use of the raster package throughout the
course where a lot of the handling is done for us, however sometimes we
need to manipulate image data with more advanced techniques and it is
therefore crucial to understand the underlying data structure.
Data Frames and Lists
Generally speaking, a data.frame is a list of vectors of
equal length, which can have varying data types.
# Create data.frame with four columns of different data types
df <- data.frame(sensor = c('Landsat-5 TM', 'Landsat-7 ETM+', 'Landsat-8 OLI-TIRS'),
n_bands = c(7, 8, 11),
active = c(F, T, T),
launched = c(1984, 1999, 2013))
print(df)## sensor n_bands active launched
## 1 Landsat-5 TM 7 FALSE 1984
## 2 Landsat-7 ETM+ 8 TRUE 1999
## 3 Landsat-8 OLI-TIRS 11 TRUE 2013The top line of the data.frame is the header, describing
the column names. Each entry is called a cell and may be indexed
(accessed) individually. We will see how this works in a moment.
Lastly, let us have a look at lists. Lists are objects that can
contain elements of different types – vectors, string scalars, matrices,
functions or yet another list. Hence, a list is a generic vector
containing other objects. The dimensions of the elements to be included
must not be identical, which gives us additional flexibility to
integrate heterogeneous datasets. A list can be created using the
list() function:
# Store the matrix, array and vector in a list
l <- list(m3, a, df)
print(l)## [[1]]
## [,1] [,2]
## [1,] 1 2
## [2,] 3 4
## [3,] 5 6
## [4,] 7 8
## [5,] 9 10
##
## [[2]]
## , , 1
##
## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 1
## [3,] 3 7 2
## [4,] 4 8 3
##
##
## [[3]]
## sensor n_bands active launched
## 1 Landsat-5 TM 7 FALSE 1984
## 2 Landsat-7 ETM+ 8 TRUE 1999
## 3 Landsat-8 OLI-TIRS 11 TRUE 2013As the print results suggests, our three objects are now
stored at locations [[1]], [[2]] and
[[3]]. We may want to access these list elements and cells
therein individually. This brings us to the next topic - indexing.
Indexing
So how do we access values in vectors,
data.frames or the elements in the list we
just created? Each element is assigned an index (i.e. a positive
integer) which we can retrieve by addressing these indices. This is
achieved using square brackets and a given index location(s).
b <- c(2, 4, 8, 16, 32, 64, 128, 256)
# Retrieve the 4th value of b
b[4]
# Retrieve value 1, 2, and 3
b[1:3] ## [1] 16
## [1] 2 4 8Use a vector to select certain positions:
# Retrieve value 1, 3, 5, and 7 of b
b[c(1, 3, 5, 7)] ## [1] 2 8 32 128Manipulate entries:
# Set the first value in b to 0
b[1] <- 0
print(b)## [1] 0 4 8 16 32 64 128 256For lists, one generally uses [[ to select any single
element, whereas [ returns a list of the selected
elements.
# [[]] returns element 1 in its original data type
typeof(l[[1]])
# [] returns a list containing element 1
typeof(l[1])
# Create a new object containing element 1
object_l <- l[[1]]
# Create a new object containing cell [4,2] of element 1
cell_l <- l[[1]][4,2] ## [1] "integer"
## [1] "list"We may also use the $ operator to access specific
elements (in lists) or columns (in data frames), given that names were
assigned. For example:
names(l) # Currently, our list does not have names...
names(l) <- c('matrix', 'array', 'df') #... names() can be used to assign element names
print(l$df)
print(df$n_bands)## NULL
## sensor n_bands active launched
## 1 Landsat-5 TM 7 FALSE 1984
## 2 Landsat-7 ETM+ 8 TRUE 1999
## 3 Landsat-8 OLI-TIRS 11 TRUE 2013
## [1] 7 8 11If you are uncertain about the characteristics of an object, you may use additional functions to investigate its attributes. Good examples are:
str(): structure of an objectlength(): number of elements in one-dimensional objects or listsnrow(),ncol(): number of rows or columns in 2-dimensional objects (df or matrix)class(),typeof(): class or type of an objectnames(): retrieve column names of df, or element names of list
Missing Values
Often our data has missing entries. For illustration, think of a
vector of temperature measurements where due to measurement error we
miss an entry now and then. For completeness, those are not just
excluded but may often be represented by a certain value, e.g. -9999. In
R missing values can be set to the logical constant NA (Not
Available).
Let us consider an vector of hourly temperature measurements where -9999 in fact was used to represent erroneous measurements:
t_hourly <- c(10.1, 8.6, 8.2, 7.2, 6.8, -9999, 8.2, 9, 9.5, 10.5, -9999, 9.9, 9.7, 8, 7.7, 7.1, 6.8, 5.8, 5.2, 4.2, 4.0, 4.1, 4.2, 3.8)
# Calculate daily mean temperature
mean(t_hourly)## [1] -826.6417These measurements exhibit two invalid temperature values of
-9999. Accordingly, the calculated mean is not of value to
us. We need to flag these entries as invalid or more generally not
available NA:
# Use indexing with logical condition
t_hourly[t_hourly == -9999] <- NA
# ...ifelse to manipulate array: ifelse(condition, TRUE, FALSE)
t_hourly <- ifelse(t_hourly == -9999, NA, t_hourly)
# mean() will yield NA as soon as one NA value appears unless...
mean(t_hourly)
# ... we specify na.rm = True to ignore missing values:
mean(t_hourly, na.rm = T)## [1] NA
## [1] 7.209091There are often various ways to achieve what we want, the best way largely depends on how we want to proceed with our data. Here are more examples to do the same:
t_hourly <- c(10.1, 8.6, 8.2, 7.2, 6.8, -9999, 8.2, 9, 9.5, 10.5, -9999, 9.9, 9.7, 8, 7.7, 7.1, 6.8, 5.8, 5.2, 4.2, 4.0, 4.1, 4.2, 3.8)
# Use only the entries that are not -9999...
mean(t_hourly[t_hourly != -9999])
# ...or use non-NAs after assigning NA to -9999
t_hourly[t_hourly == -9999] <- NA
mean(t_hourly[!is.na(t_hourly)])## [1] 7.209091
## [1] 7.209091Flow Control
Often we want code to be executed only when certain conditions are
met (conditional statements) or repeat a certain chain of commands in
order (repeating operations). For conditional statements, the most
commonly used approach are if statements which are based on
boolean logic. Accordingly, it is evaluated if a condition is
TRUE or FALSE and the following code block
will therefore be executed (TRUE) or not
(FALSE).
if-statements
The basic form of an if statement could be as
follows:
# Reclassify an image based on a certain NDVI threshold, after a sanity check if the threshold is valid
ndvi_threshold <- 0.8
if (ndvi_threshold > 1 | ndvi_threshold < -1) { # i.e. if NDVI larger 1 OR smaller -1
print('Invalid NDVI threshold provided. Must be in range -1 to 1.')
} else {
print('Valid NDVI threshold. Continue processing ...')
# Additional code to be executed follows here ....
}## [1] "Valid NDVI threshold. Continue processing ..."Above we already used the vectorized version ifelse()
which for instance allowed us to manipulate out vector of temperature
records:
t_hourly <- ifelse(t_hourly == -9999, NA, t_hourly) # ifelse(condition, TRUE, FALSE)Loops
for loops can be used to iterate over items and
repeatingly execute code blocks:
# Generate sequence of integers from 1 to 10 with a step size of 1
v <- seq(1, 10, 1)
# Iterate over each entry in v and print
for (item in v){
print(item)
}## [1] 1
## [1] 2
## [1] 3
## [1] 4
## [1] 5
## [1] 6
## [1] 7
## [1] 8
## [1] 9
## [1] 10while repeatedly evaluates a condition and executes
commands until the final condition is fulfilled.
# Assign integer to v
v <- -3
# Adds +1 to v and then prints until v > 2
while (v <= 2) {
v <- v + 1
print(v)
}## [1] -2
## [1] -1
## [1] 0
## [1] 1
## [1] 2
## [1] 3Note that loops should be avoided in higher-level languages such as R or Python where possible because they tend to make the programs run slower than when using in-built vectorized functions. For example, calculating the sum over a vector may be calculated as follows:
s <- seq(1, 10, 0.5)
s_sum <- 0
for (i in s){
s_sum <- s_sum + i
}
print(s_sum)## [1] 104.5Much more elegant and faster (think of manipulating large datasets) is to use
s_sum <- sum(s)
print(s_sum)## [1] 104.5Functions
R allows for user-specified functions which comes in handy whenever a certain computation needs to be repeated or generalized. Generally, a function is a set of statements organized together to perform a specific task. Using functions greatly enhances code structure as it improves the clarity and readability of your script.
Function arguments are provided in round () brackets.
Between {} stands the function body which contains the
collection of statements. return() is used to define the
output of a function. A function may be as simple as a print statement
without any objects to be returned.
A function object is defined with the function()
call:
# Basic function
printer <- function(){
print('Hello World!')
}
printer()## [1] "Hello World!"Another example where we want the function to calculate the NDVI for a given pair of reflectance values in the red and near-infrared region of the spectrum:
# Function to calculate the NDVI
calc.ndvi <- function(red, nir, scale_factor=1){
ndvi <- (nir-red)/(nir+red)
return(ndvi*scale_factor)
}
p_red <- 0.2
p_nir <- 0.8
ndvi <- calc.ndvi(p_red, p_nir)
print(ndvi)## [1] 0.6Here, red and nir are obligatory arguments
that the user has to specify while scale_factor is optional
as such that it has a default value which may or may not be altered.
Tutorials
We here covered only very basic aspects of R. At this stage, we did
not cover advanced data wrangling using packages such as
tidyr or dplyr, or look into advanced plotting
with ggplot2. Please follow one or several tutorials to
revisit and amend your knowledge:
General tutorials:
Graphics & visualization:
Geodata processing:
- Wageningen University: Geoscripting
- University of Colorado: EarthDataScience)
- RSpatial: Remote Sensing Image Analysis
Coding style
A few basic rules apply to coding in R. Here is a short summary of Hadley Wickham´s style guide:
Regularly save your progress.
Script names should be meaningful and end in ‘.R’.
Comment (#) your code & separate it into readable chunks.
Try to limit your code to 80 characters per line.
Variable and function names should be lowercase.
Variable names should be nouns and function names verbs.
Place spaces around operators (=, +, -, <-, etc.) and after commas.
Use <-, not =, for assignment.
An example:
######################################################
# Creating random data and a correlated response
# Author, 2020
# Load all required packages
library(ggplot2)## Warning: package 'ggplot2' was built under R version 4.1.2##
## Attaching package: 'ggplot2'## The following object is masked from 'package:terra':
##
## arrow# Create random data
x <- runif(50, 0, 2)
# Build function to simulate response
create.response <- function(x){x + rnorm(50, 0, 0.2)}
# Apply function to random data
y <- create.response(x)
# Make a dataframe
data <- data.frame('x' = x, 'y' = y)
# Plot the simulated dataset
ggplot(data, aes(x = x, y = y)) +
geom_point()
# Investigate correlation in the data
cor(data$x, data$y)## [1] 0.9546448Help!
If you get stuck while programming, there are plenty of things you can do:
- Seek the function´s help page (i.e. highlight the function and hit F1)
- Search your problem or error message
- Ask your colleagues
- Use the moodle course forum
- Check forums (e.g., StackOverflow)
Copyright © 2020 Humboldt-Universität zu Berlin. Department of Geography.