- R packages
- Install and load
- Namespace
- R base graphics
- Plots
- Histogram
- Scatterplot
- Boxplot
ggplot2- Histogram
- Scatterplot
- Boxplot
- Grouping by factor
- Scaling by numeric variables
- Trellis graphs
- Combining multiple layers
- Axis text and plot title
- Multi-plot graphics
- Graphics file formats
- Data manipulation
- Tidy data
- pivot_longer() and pivot_wider()
- separate()
- Pipes
- Mutating new variables
- Creating summaries
- Grouping data
- Tibbles
- Subsetting data
- Reading
3. Visualization and Manipulation
Humboldt-Universität zu Berlin | Geography Department
R packages
Install and load
A package is a collection of previously programmed functions, often including functions for specific tasks. It is tempting to call this a library, but the R community refers to it as a package.
There are two types of packages: those that come with the base
installation of R and packages that you must manually download and
install using install.packages("package"). You can load a
package with library(package).
One very powerful graphics package is ggplot2.
install.packages("ggplot2")You can keep your packages up to date with with the following command:
update.packages()You only need to install a package once, but you need to load it every time you start a new R-Session!
library(ggplot2) # visualization
library(gridExtra) # visualization
library(tidyr) # manipulating data (tidy messy data)
library(dplyr) # manipulating dataNamespace
Sometimes functions from different packages have a common name (e.g.,
select() in the raster and
dplyr packages). We therefore need to tell R which
function it should use by defining the namespace for the function:
raster::select()
dplyr::select()R base graphics
Plots
R’s base graphics are great for quick visualization of
your data. The main base graphics functions are plot(),
hist() and boxplot():
Histogram
hist(airquality[ , c("Ozone")])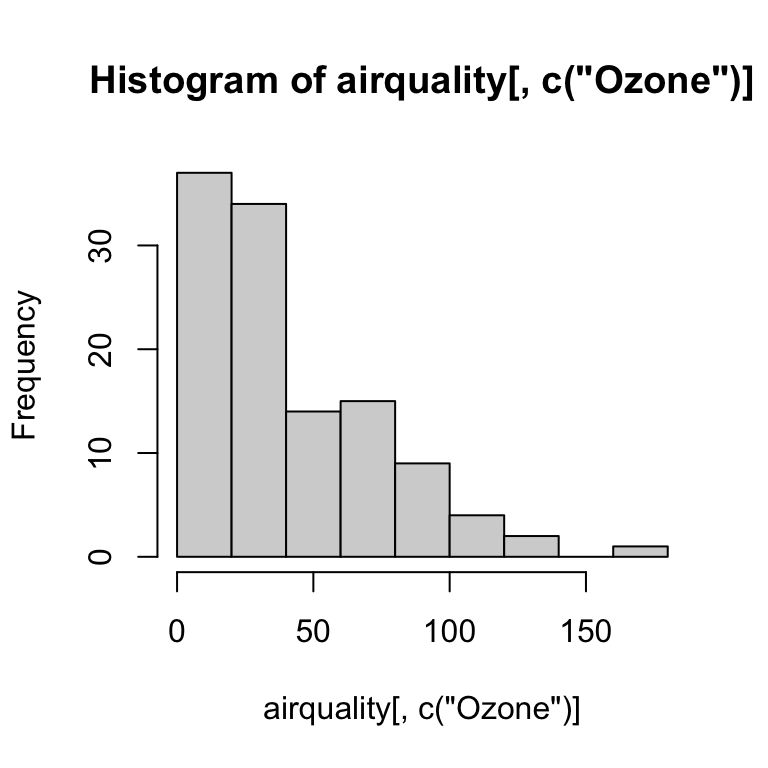
Scatterplot
plot(airquality[ , c("Ozone")], airquality[ , c("Temp")])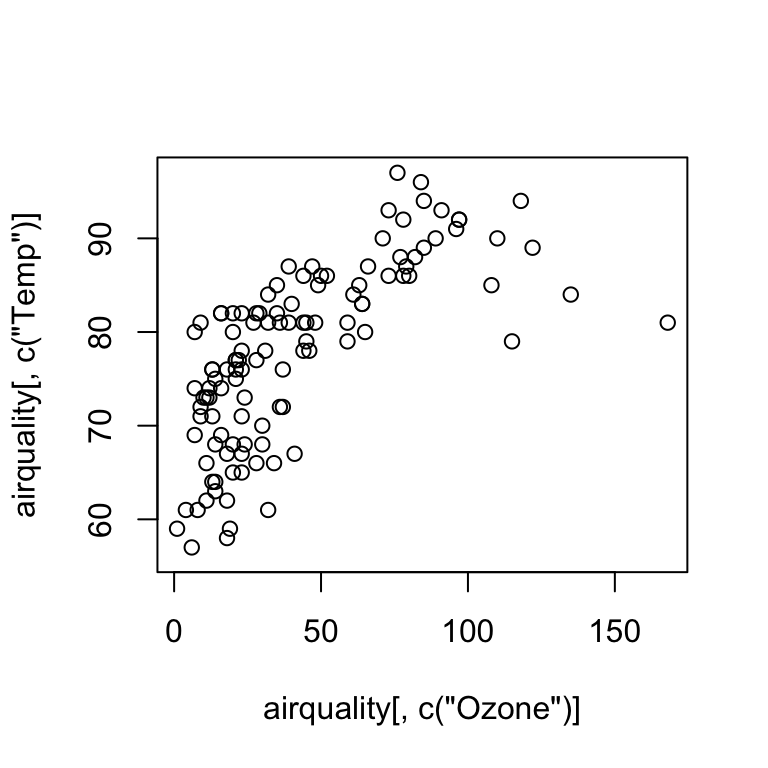
Boxplot
boxplot(airquality[ , c("Ozone")] ~ airquality[ , c("Month")])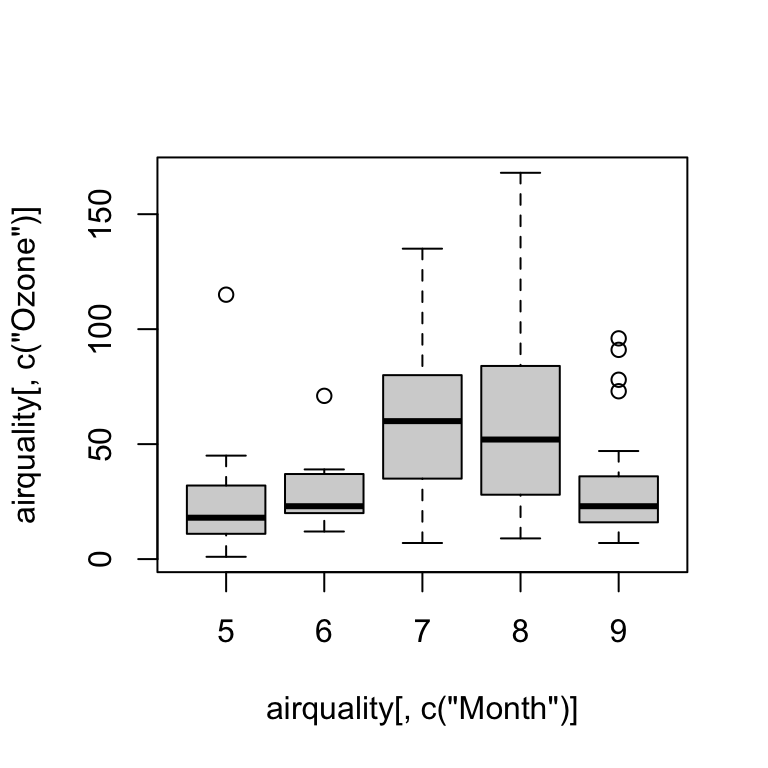
ggplot2
The base graphics system can also be used visualize complex datasets
and relationships, but you will need to write more complex code and
functions. The ggplot2 package is a great alternative, but
it takes some getting used to. It works somewhat differently than base R
and graphics.
When you create a ggplot, you have to define at least three key components:
dataattribute in theggplot()orgeom_*()function call specifies the data set used for the entire plot or individual geom_ layers, respectively.aesthetics: function
aes()is used to map (link) variable names in the data to the axes (e.g., x and y) and visual properties of the graph (e.g., color and shape of symbols).at least one layer which describes how to render each observation. Layers are usually created with a geom function, e.g.
geom_histogram(),geom_point(),geom_boxplot(), and many more.
Histogram
ggplot(data = airquality, aes(x = Ozone)) +
geom_histogram()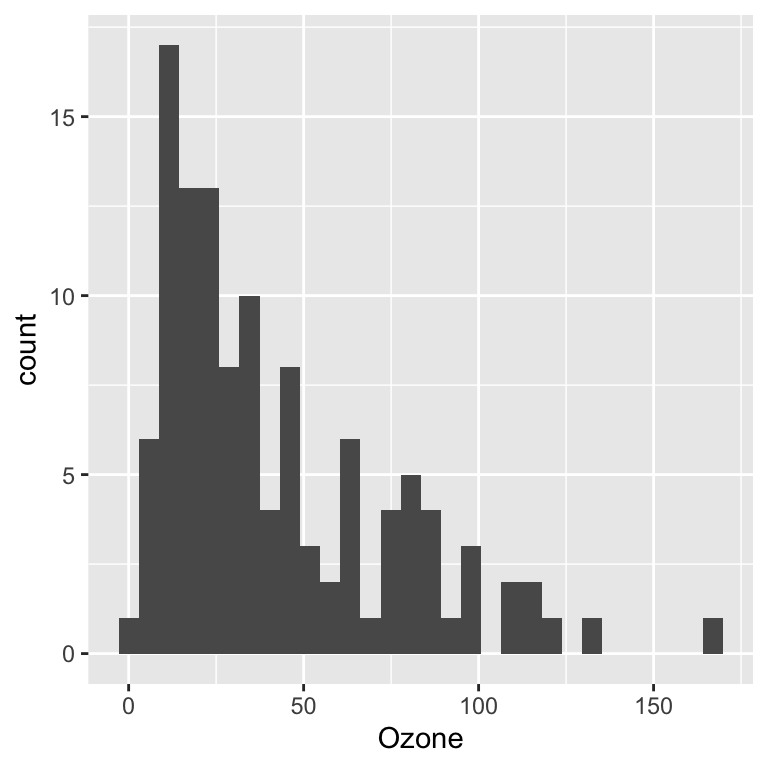
Scatterplot
ggplot(data = airquality, aes(x = Ozone, y = Temp)) +
geom_point()
Boxplot
ggplot(data = airquality, aes(x = factor(Month), y = Ozone)) +
geom_boxplot()
Grouping by factor
You can use factor variables to change the
aesthetics of geom_point and
geom_line layers. The following example visualizes the data
from different months with different colors.
ggplot(data = airquality, aes(x = Ozone, y = Temp, col = factor(Month))) +
geom_point()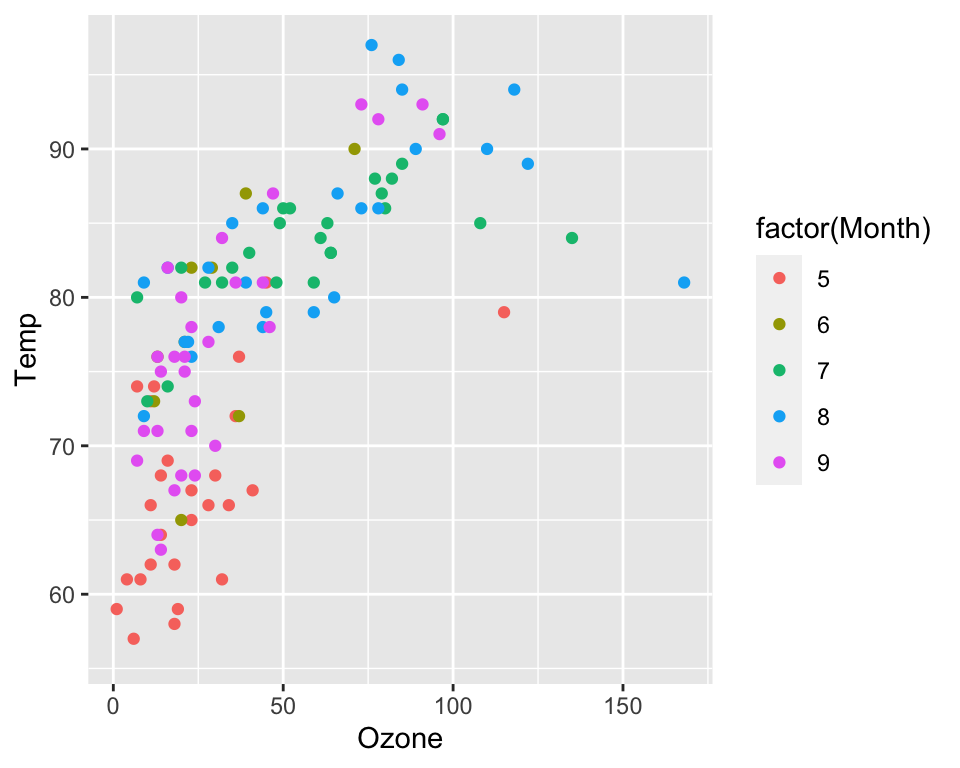
You can also display factors through different symbols (with
geom_point):
ggplot(data = airquality, aes(x = Ozone, y = Temp, shape = factor(Month))) +
geom_point()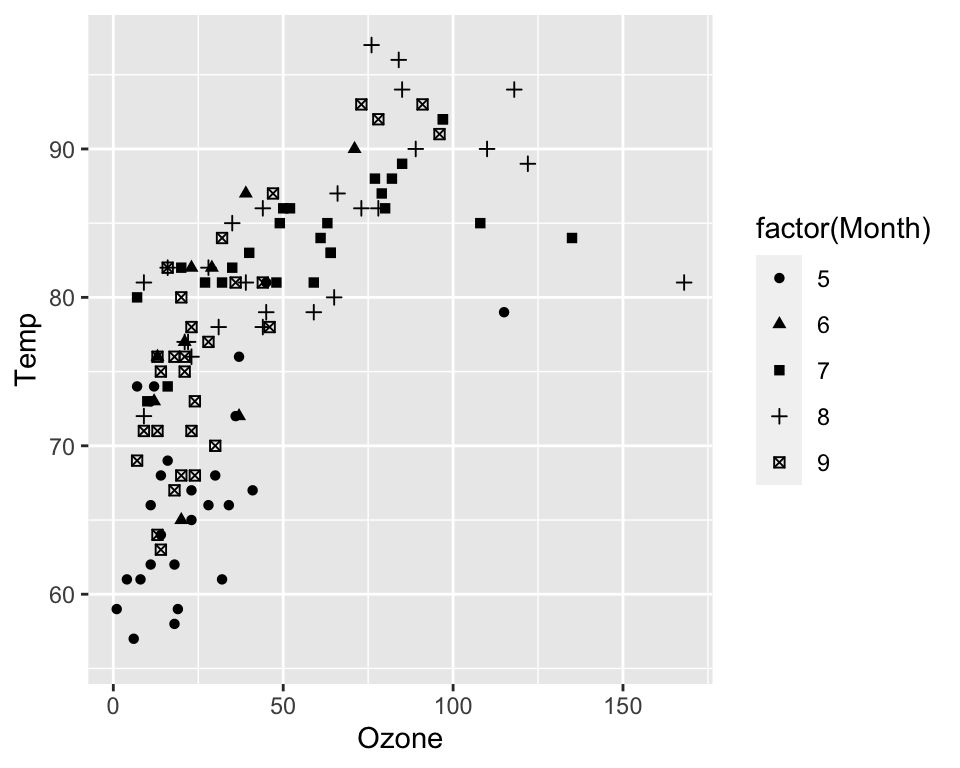
…or the line type with geom_line:
ggplot(data = airquality, aes(x = Day, y = Temp, linetype = factor(Month))) +
geom_line()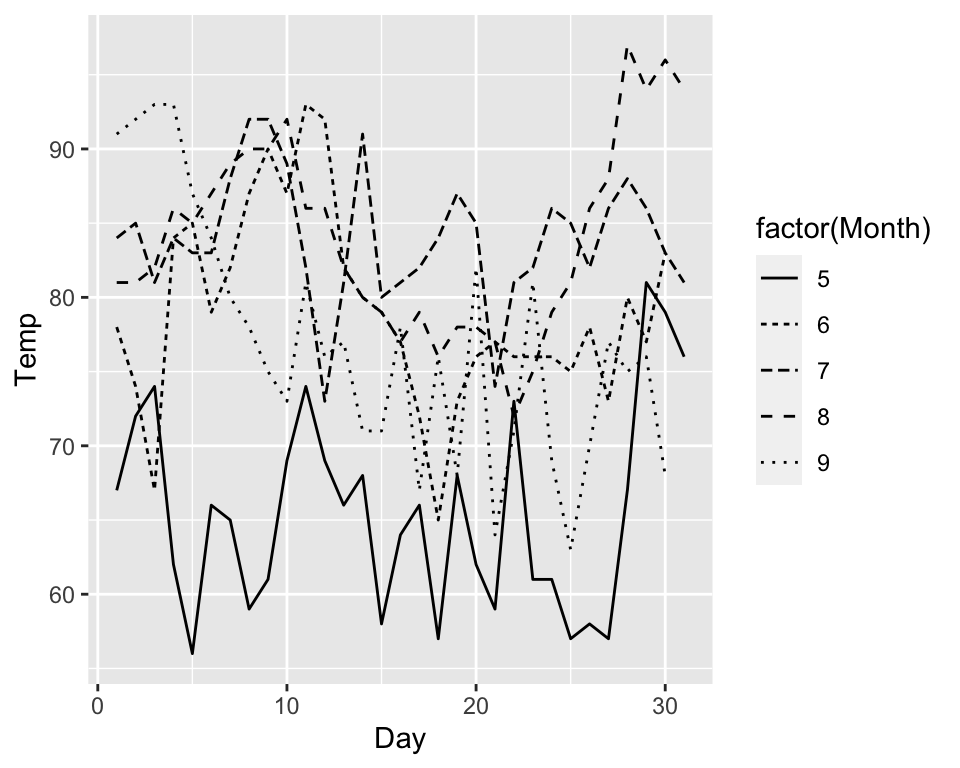
..or the fill color of geom_boxplot and
geom_histogram:
ggplot(data = airquality, aes(x = Ozone, fill = factor(Month))) +
geom_histogram()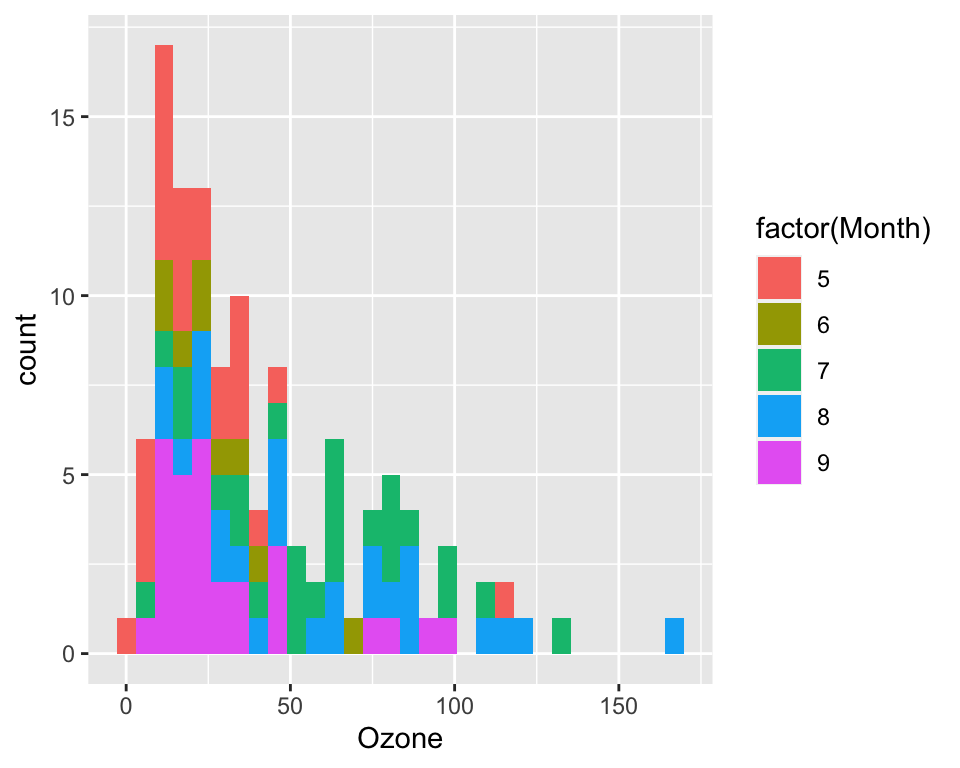
Scaling by numeric variables
Factors can be used to define colors and symbols. Numeric varibles can be displayed by varying symbol sizes and color gradients:
ggplot(data = airquality, aes(x = Ozone, y = Temp, col = Wind, size = Solar.R)) +
geom_point()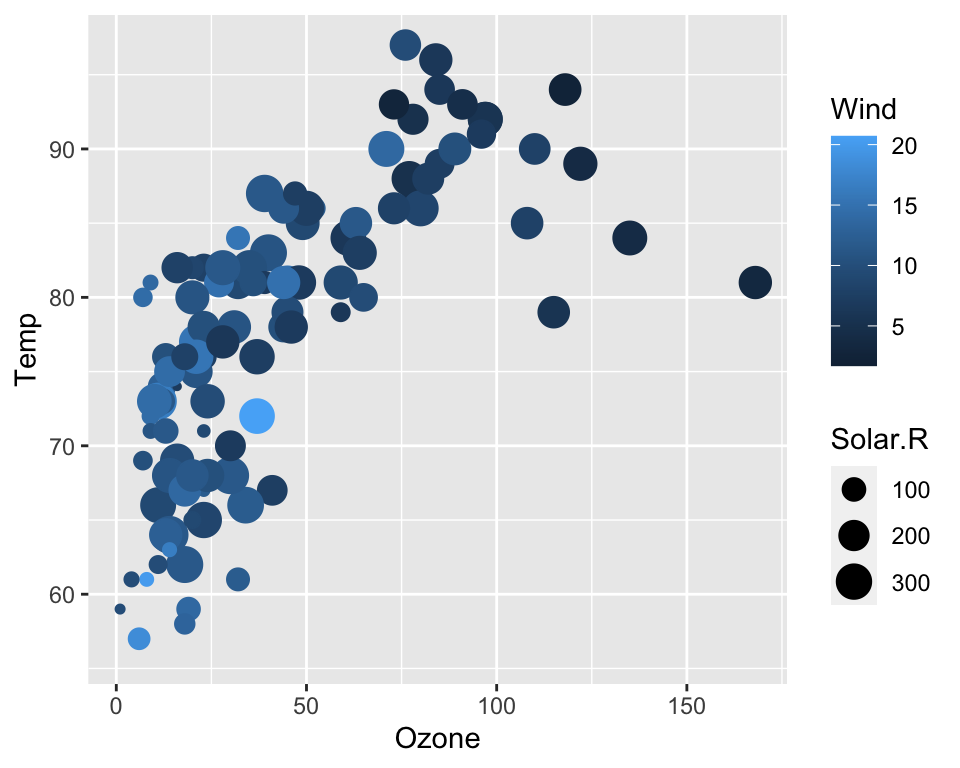
Trellis graphs
Trellis plots are based on the idea of conditioning on the values taken on by one or more of the variables in a data set.
In the case of a categorical variable, this means carrying out the same plot for the data subsets corresponding to each of the levels of that variable.
R provides built-in trellis graphics with the lattice
package.
In ggplot2 trellis plots (here called multi-facet plots)
can be achieved with facet_wrap():
ggplot(data = airquality, aes(x = Ozone, y = Temp)) +
geom_point() +
facet_wrap(~Month)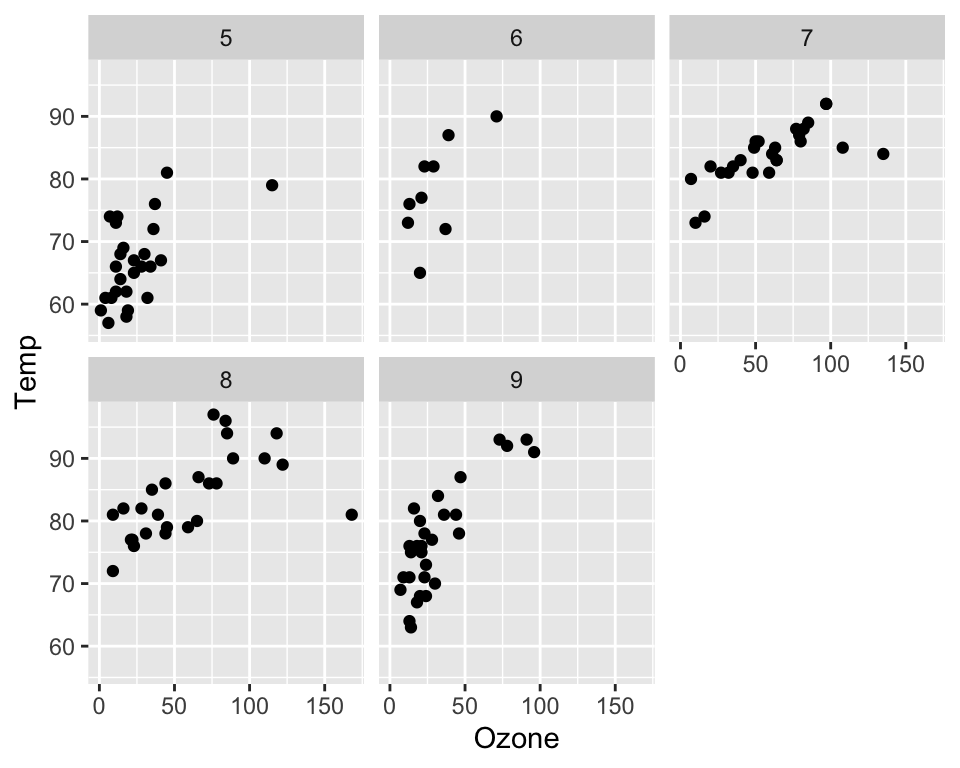
The axes among individual plots (pannels) can be either fixed or variable (free):
ggplot(data = airquality, aes(x = Ozone, y = Temp)) +
geom_point() +
facet_wrap(~Month, scales = "free")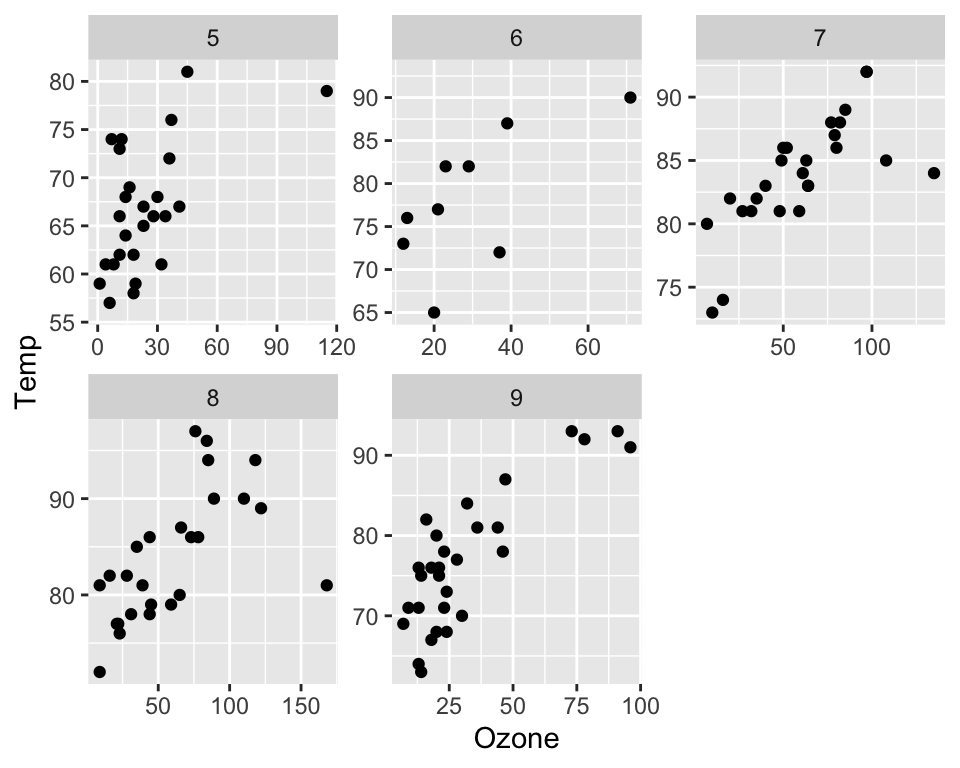
Combining multiple layers
A ggplot can contain multiple layers and each layer can have its own
aesthetics:
ggplot(data = airquality, aes(x = Day, y = Temp)) +
geom_point(aes(size = Ozone)) +
geom_line(aes(linetype = factor(Month)))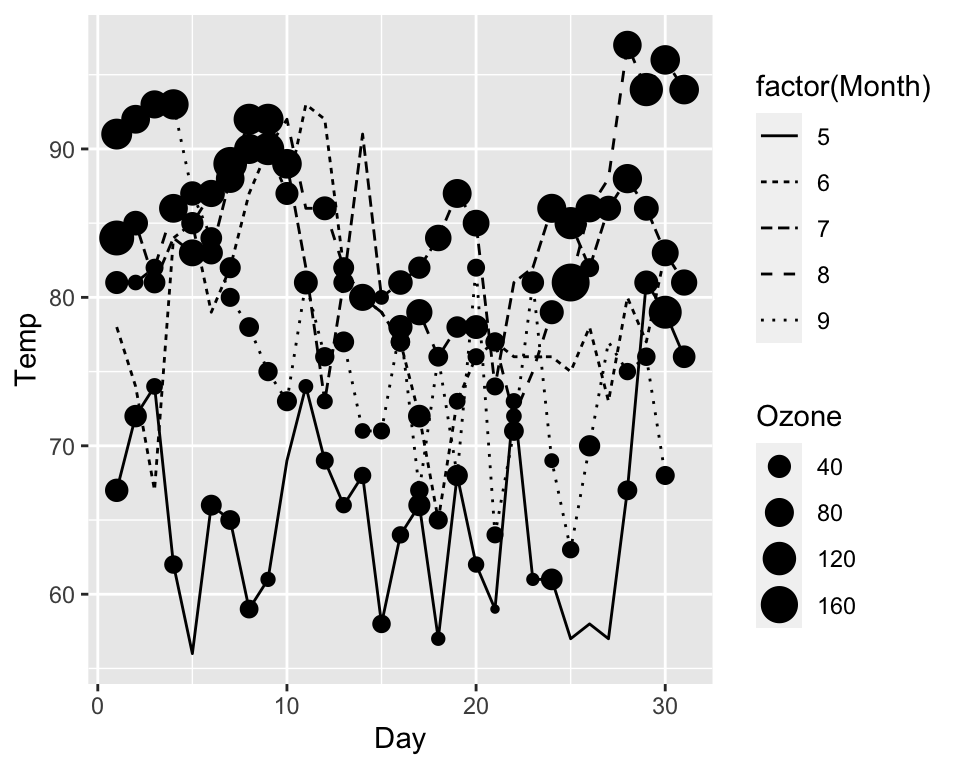
Axis text and plot title
Modify axes titles with xlab() and ylab(),
and plot title with ggtitle():
ggplot(data = airquality, aes(x = Ozone, y = Temp, col = factor(Month))) +
geom_point() +
labs(x = "Ozone concentration [ppm]",
y = "Temperature [°F]",
title = "Relationship between Ozone und Temperture")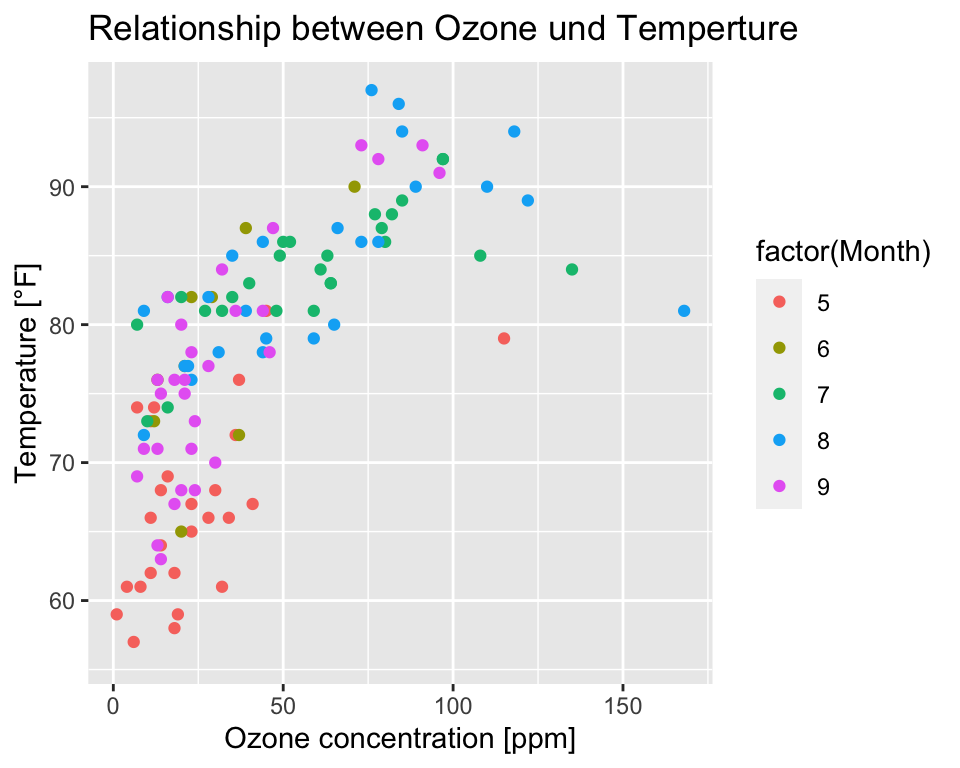
Multi-plot graphics
You can combine multiple plots in a single graphic layout with
grid.arrange() of the gridExtra package.
First, you must create a graphics object for each plot. Then the two
objects are combined with grid.arrange().
plot1 <- ggplot(data = airquality, aes(x = Ozone, y = Temp)) + geom_point()
plot2 <- ggplot(data = airquality, aes(x = Ozone, y = Wind)) + geom_point()
gridExtra::grid.arrange(plot1, plot2, ncol = 2)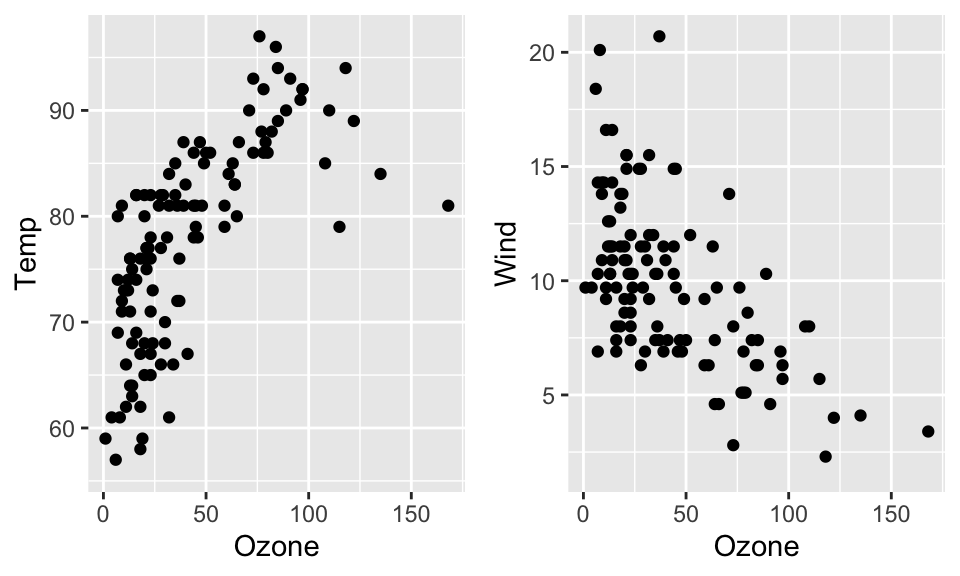
Graphics file formats
R graphics can be exported into a number of file formats:
- tiff
- png
- jpg
- …
To export graphics you need to ‘open’ a new Graphic
Device before running your plot functions and close it when you
are done. The ‘opening’ function depends on the format
(e.g. pdf(), tif(), png(), etc.).
To close the graphics device you need to call
dev.off().
pdf(file = "Figures/scatterplot01.pdf", width = 7, height = 5)
ggplot(data = airquality, aes(x = Ozone, y = Temp, col = factor(Month))) +
geom_point()
dev.off()Alternatively, you can use the ggsave() function.
p <- ggplot(data = airquality, aes(x = Ozone, y = Temp, col = factor(Month))) +
geom_point()
ggsave(filename = "plot.pdf", plot = p, path = "Figures")Data manipulation
Tidy data
Cleaning and preparing (tidying) data for analysis can make up a substantial proportion of the time spent on a project. It is therefore good practice to follow certain guidelines for structuring your data (Wickam, 2014):
- Each variable forms a column.
- Each observation forms a row.
- Each type of observational unit forms a table.
Real datasets can, and often do, violate the three precepts of tidy data in almost every way imaginable. While occasionally you do get a dataset that you can start analysing immediately, this is the exception, not the rule. This section describes the five most common problems with messy datasets, along with their remedies:
- Column headers are values, not variable names.
- Multiple variables are stored in one column.
- Variables are stored in both rows and columns.
- Multiple types of observational units are stored in the same table.
- A single observational unit is stored in multiple tables.
Most messy datasets, including types of messiness not explicitly described above, can be tidied with a small set of tools.
Basically, data is often recorded in this format (wide-format):
| Plot | Measure1 | Measure2 |
|---|---|---|
| 1 | 13.1 | 12.7 |
| 2 | 9.5 | 10.6 |
| 3 | 11.9 | 7.4 |
But we want to have it in following format (long-format)
| Plot | Measure | Value |
|---|---|---|
| 1 | M1 | 13.1 |
| 2 | M1 | 9.5 |
| 3 | M1 | 11.9 |
| 1 | M2 | 12.7 |
| 2 | M2 | 10.6 |
| 3 | M2 | 7.4 |
pivot_longer() and pivot_wider()
yields <- read.table("data/yields.txt", sep = ";", header = TRUE)
yields[1:5, ]## plotid sand clay loam
## 1 1 6 17 13
## 2 2 10 15 16
## 3 3 8 3 9
## 4 4 6 11 12
## 5 5 14 14 15The pivot_longer() function in the
tidyr package can be used to convert the previous
dataset into a tidy dataset. In the example below, we want to
reshape the sand, clay, and loam
columns, hence we exclude the plotid column (denoted with a
minus sign). See also here, for
ways to select columns in the tidyverse.
yields_tidy <- tidyr::pivot_longer(yields, cols= -plotid, names_to = "soiltype", values_to = "yield")
# pivot_longer(yields, cols= starts_with(c("sand", "loam", "clay")), names_to = "soiltype", values_to = "yield")
yields_tidy[c(1:3, 11:13), ]## # A tibble: 6 × 3
## plotid soiltype yield
## <int> <chr> <int>
## 1 1 sand 6
## 2 1 clay 17
## 3 1 loam 13
## 4 4 clay 11
## 5 4 loam 12
## 6 5 sand 14The dataset is now tidy, i.e. ready for analysis and plotting:
ggplot(yields_tidy, aes(x = soiltype, y = yield)) +
geom_boxplot()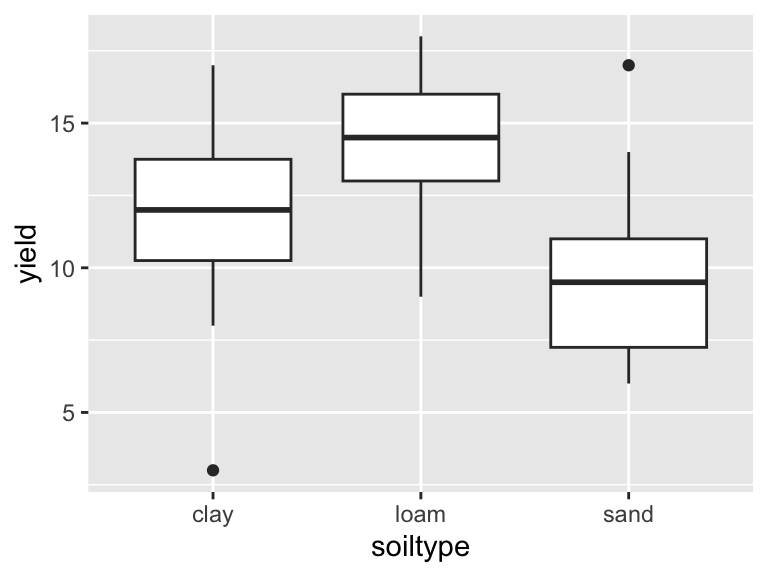
Sometimes we want to back-transform the data into wide format, which
can be done using the pivot_wider() function:
yields <- pivot_wider(yields_tidy, id_cols=plotid, names_from=soiltype, values_from=yield)
yields[1:3, ]## # A tibble: 3 × 4
## plotid sand clay loam
## <int> <int> <int> <int>
## 1 1 6 17 13
## 2 2 10 15 16
## 3 3 8 3 9Sometimes column names include several attributes:
yields2 <- read.table("data/yields2.txt", sep = ";", header = TRUE)
yields2[1:3, ]## plotid sand_fine clay_fine loam_fine sand_coarse clay_coarse loam_coarse
## 1 1 6 17 13 9 18 15
## 2 2 10 15 16 10 15 18
## 3 3 8 3 9 9 3 9After transforming it into long format, the data still is not a tidy data set (‘Multiple variables are stored in one column’):
yields2_tidy <- pivot_longer(yields2, cols= starts_with(c("sand", "loam", "clay")), names_to = "soiltype", values_to = "yield")
yields2_tidy[c(1:3, 11:13), ]## # A tibble: 6 × 3
## plotid soiltype yield
## <int> <chr> <int>
## 1 1 sand_fine 6
## 2 1 sand_coarse 9
## 3 1 loam_fine 13
## 4 2 clay_fine 15
## 5 2 clay_coarse 15
## 6 3 sand_fine 8separate()
We need to split the soiltype column into two columns
soildtype and grain. This can easily be done
using the separate() function:
yields2_tidy <- separate(yields2_tidy, col = soiltype, into = c("soiltype", "grain"), sep = "_")
yields2_tidy[c(1:3, 11:13), ]## # A tibble: 6 × 4
## plotid soiltype grain yield
## <int> <chr> <chr> <int>
## 1 1 sand fine 6
## 2 1 sand coarse 9
## 3 1 loam fine 13
## 4 2 clay fine 15
## 5 2 clay coarse 15
## 6 3 sand fine 8Which can now be nicely plotted using ggplot2:
ggplot(yields2_tidy, aes(x = soiltype, y = yield, fill = grain)) +
geom_boxplot()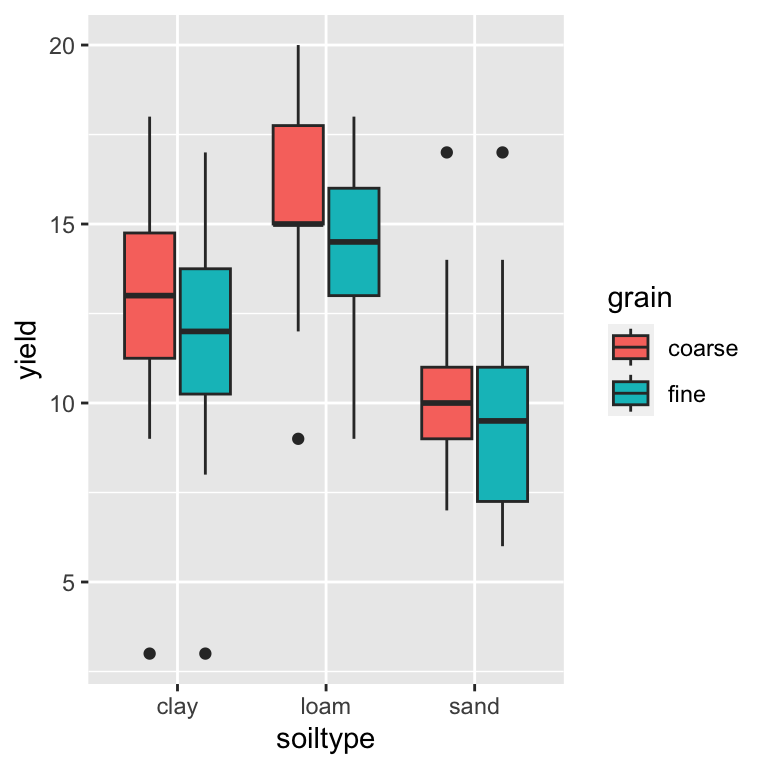
Pipes
Often we have to do many steps to wrangle our data into a tidy form, which can become messy and un-readable. This is especially true if we nest many steps into one line. Using so-called pipes in the dplyr package can help us:
yields2_tidy <- yields2 %>%
pivot_longer(., cols= -plotid, names_to = "soiltype", values_to = "yield") %>%
separate(., col = soiltype, into = c("soiltype", "grain"), sep = "_")The term x <- y %>% f1(.) %>% f2(.) is similar
to x <- f2(f1(y)), but more readable when the the
statement is long!
Mutating new variables
If we want to create a new variable (column), we can use the
mutate() function:
yields2_tidy <- yields2_tidy %>%
mutate(yield_log = log(yield),
yield_log10 = log10(yield))
yields2_tidy[1:3, ]## # A tibble: 3 × 6
## plotid soiltype grain yield yield_log yield_log10
## <int> <chr> <chr> <int> <dbl> <dbl>
## 1 1 sand fine 6 1.79 0.778
## 2 1 clay fine 17 2.83 1.23
## 3 1 loam fine 13 2.56 1.11Creating summaries
If we want to create a summary of (a) variable(s), we can use the
summarize() function:
yields2_tidy_summary <- yields2_tidy %>%
summarize(yield_mean = mean(yield),
yield_median = median(yield))
yields2_tidy_summary## # A tibble: 1 × 2
## yield_mean yield_median
## <dbl> <dbl>
## 1 12.4 12Grouping data
Using dyplr we can easily calculate new variables or derive summaries of our data. However, often we want to do both wihtin specific groups, e.g. the mean/median yield within each soil-type and grain-class. We therefore need to group the data:
yields2_tidy_summary <- yields2_tidy %>%
group_by(soiltype, grain) %>%
summarize(yield_mean = mean(yield),
yield_median = median(yield))## `summarise()` has grouped output by 'soiltype'. You can override using the
## `.groups` argument.yields2_tidy_summary## # A tibble: 6 × 4
## # Groups: soiltype [3]
## soiltype grain yield_mean yield_median
## <chr> <chr> <dbl> <dbl>
## 1 clay coarse 12.4 13
## 2 clay fine 11.5 12
## 3 loam coarse 15.5 15
## 4 loam fine 14.3 14.5
## 5 sand coarse 10.6 10
## 6 sand fine 9.9 9.5We also can use the mutate() function within groups,
e.g. for centering the yields within groups:
yields2_tidy <- yields2_tidy %>%
group_by(soiltype, grain) %>%
mutate(yield_centered = yield - mean(yield))
yields2_tidy## # A tibble: 60 × 7
## # Groups: soiltype, grain [6]
## plotid soiltype grain yield yield_log yield_log10 yield_centered
## <int> <chr> <chr> <int> <dbl> <dbl> <dbl>
## 1 1 sand fine 6 1.79 0.778 -3.9
## 2 1 clay fine 17 2.83 1.23 5.5
## 3 1 loam fine 13 2.56 1.11 -1.3
## 4 1 sand coarse 9 2.20 0.954 -1.6
## 5 1 clay coarse 18 2.89 1.26 5.6
## 6 1 loam coarse 15 2.71 1.18 -0.5
## 7 2 sand fine 10 2.30 1 0.100
## 8 2 clay fine 15 2.71 1.18 3.5
## 9 2 loam fine 16 2.77 1.20 1.7
## 10 2 sand coarse 10 2.30 1 -0.600
## # ℹ 50 more rowsTibbles
Note: The result from a dplyr call is a so-called tibble, which is similar to a data frame, but has some improvements. For example, it shows the data type and dimensions of the data frame, as well as it is easier to print in the console.
head(yields_tidy)## # A tibble: 6 × 3
## plotid soiltype yield
## <int> <chr> <int>
## 1 1 sand 6
## 2 1 clay 17
## 3 1 loam 13
## 4 2 sand 10
## 5 2 clay 15
## 6 2 loam 16Subsetting data
Often, we also want to exclude observations (rows), which we can
easily do using the filter() function:
yields2_tidy_subset <- yields2_tidy %>%
filter(grain == "fine")Similarly, we can exclude variables (columns) that we don’t need for
further analysis using the select() function:
yields2_tidy_subset <- yields2_tidy %>%
select(-yield_log10, -yield_log)Thereby we ca either select columns or drop them using a ‘-’ before the columns name.
Reading
Wickam, H. 2014. Tidy data. Journal of Statistical Software, 59, 1-23. doi:10.18637/jss.v059.i10
RStudio 2015. Data Wrangling with dplyr and tidyr Cheat Sheet
Copyright © 2024 Humboldt-Universität zu Berlin. Department of Geography.