- Overview
- Packages
- Data
- Meuse points
- Meuse grid
- Create a grid
- IDW interpolation
- Stationarity
- Lagged scatter plots
- Variogram cloud
- Sample variogram
- Direction dependence
- Cutoff and lag width
- Theoretical variogram
- Fit a variogram
- Partial variogram model fitting
- Visual variogram model fitting
- Ordinary kriging
- Universal kriging
- Comparison
- References
Spatial interpolation in R
Humboldt-Universität zu Berlin | Geography Department
Overview
- Inverse Distance Weighted interpolation
- Semivariogram
- Kriging
Packages
We will mostly deal with package gstat, because it
offers the widest functionality in the geostatistics curriculum for R:
it covers variogram cloud diagnostics, variogram modeling, everything
from global simple kriging to local universal cokriging, multivariate
geostatistics, block kriging, indicator and Gaussian conditional
simulation, and many combinations.
The functions in gstat take spatial vector and raster
data as input. For spatial vector data, we already introduced the
sf package (see this
lab). In terms of raster data, gstat only understands
formats from the older sp package and the newer
stars package. All are from the same developers. The
stars package was build to read and manipulate
spatio-temporal array data (Pebesma, 2020). So this
includes the usual raster data like satellite images and climate grids,
but compared to other raster data packages, it extents the interface to
incorporate the temporal dimension. Specifically, stars objects can have
a time dimension in addition to the spatial 2D dimensions (longitude and
latitude), where the time dimension can take Date
values.
library("gstat") # geostatistics
library("mapview") # map plot
library("sf") # spatial vector data
library("stars") # spatial-temporal data
library("terra") # raster data handling
library("ggplot2") # plotting
mapviewOptions(fgb = FALSE)Data
For this tutorial, we use a dataset from Pebesma
(2022). The dataset meuse is a
tabular dataset (geopackage) that includes measurements of four heavy
metals sampled from the top soil in a flood plain along the river Meuse.
The dataset also contains a number of covariates. We also have a raster
of some of the covariates meuse_gridcv. Further, we learn
how to create our own grid for the sampled area: meuse_grid.
Meuse points
Let’s read the geopackage of the soil samples using the
read_sf() function of the sf package (see this lab).
meuse <- read_sf('data/meuse.gpkg')
head(meuse)## Simple feature collection with 6 features and 12 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 181025 ymin: 333260 xmax: 181390 ymax: 333611
## Projected CRS: Double_Stereographic
## # A tibble: 6 × 13
## cadmium copper lead zinc elev dist om ffreq soil lime landuse dist_m
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl>
## 1 11.7 85 299 1022 7.91 0.00136 13.6 1 1 1 Ah 50
## 2 8.6 81 277 1141 6.98 0.0122 14 1 1 1 Ah 30
## 3 6.5 68 199 640 7.8 0.103 13 1 1 1 Ah 150
## 4 2.6 81 116 257 7.66 0.190 8 1 2 0 Ga 270
## 5 2.8 48 117 269 7.48 0.277 8.7 1 2 0 Ah 380
## 6 3 61 137 281 7.79 0.364 7.8 1 2 0 Ga 470
## # ℹ 1 more variable: geom <POINT [m]>Spatial exploratory data analysis starts with the plotting of maps with a measured variable. Zinc concentration seems to be larger close to the river Meuse banks and in areas with low elevation.
mapview(meuse['zinc'])Meuse grid
The meuse grid is a geotiff raster that contains five bands: 1)
dist (distance to the Meuse river), 2) ffreq
(flooding frequency class), 3) soil type, 4)
part.a, and 5) part.b. The later two are
arbitrary divisions of the area in two areas. We can read the grid using
the read_stars() function.
meuse_gridcv <- read_stars("data/meuse_grid.tif")
meuse_gridcv## stars object with 3 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## meuse_grid.tif 0 0.1192915 1 0.9884603 1 3 25045
## dimension(s):
## from to offset delta refsys point values x/y
## x 1 78 178440 40 Double_Stereographic FALSE NULL [x]
## y 1 104 333760 -40 Double_Stereographic FALSE NULL [y]
## band 1 5 NA NA NA NA part.a,...,ffreqThe resulting stars object now contains three dimension, where the
bands are stored in the third dimension (here names band).
To be able to use this grid in gstat, we need to split the
bands into separate attributes:
meuse_gridcv <- split(meuse_gridcv, "band")
meuse_gridcv## stars object with 2 dimensions and 5 attributes
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## part.a 0 0.0000000 0.000000 0.3986465 1.000000 1.000000 5009
## part.b 0 0.0000000 1.000000 0.6013535 1.000000 1.000000 5009
## dist 0 0.1192915 0.271535 0.2971195 0.440159 0.992607 5009
## soil 1 1.0000000 1.000000 1.5775056 2.000000 3.000000 5009
## ffreq 1 1.0000000 2.000000 2.0676764 3.000000 3.000000 5009
## dimension(s):
## from to offset delta refsys point x/y
## x 1 78 178440 40 Double_Stereographic FALSE [x]
## y 1 104 333760 -40 Double_Stereographic FALSE [y]Create a grid
Spatial interpolation is done on a regular grid. The routines in
gstat require you to specify such a grid as a
stars object (or the older sp object). If you
have an existing raster dataset, you can use that as a template. Perhaps
that raster dataset also contains covariates that you want to include in
the modeling like meuse_gridcv.
Alternatively, you can create your own grids with varying spacings
between cells and a specified extent.
There are different ways to create a regular grid in R. The
sf package also includes useful functions like
st_make_grid(). Here, we use a simpler way that does not
require sf. Let’s start by grabing the bounding box
coordinates of the soil samples (meuse) to define the extent of the output
grid.
bbox <- st_bbox(meuse)
bbox## xmin ymin xmax ymax
## 178605 329714 181390 333611We can access the elements of bbox by name,
e.g. bbox$xmin. With the seq() function, we
can then create a sequence of regularly spaced coordinates in the
x and y direction. Here, I use an increment of
40, which defines the cell size.
cell_size <- 40
x <- seq(bbox$xmin, bbox$xmax, by=cell_size)
y <- seq(bbox$ymin, bbox$ymax, by=cell_size)With expand.grid(), we can combine every x
coordinate with every y coordinate. This yields a regular
grid of coordinates. The output of the function is a data.frame with the
expanded columns x and y columns.
meuse_grid <- expand.grid(x=x, y=y)
plot(meuse_grid$x, meuse_grid$y, pch=19, cex=0.1)
Now, we can convert the data.frame containing the grid coordinates to
a stars object using st_as_stars(). But first,
I add a column tmp with some fake value. Otherwise the
stars object would have 0 attributes and that would fail
the interpolation functions.
meuse_grid$tmp <- 1
meuse_grid <- st_as_stars(meuse_grid, crs=st_crs(meuse))
st_crs(meuse_grid) <- st_crs(meuse) # re-assign crs to be safeIDW interpolation
Inverse distance-based weighted interpolation (IDW) computes a weighted average,
ˆZ(s0)=∑ni=1w(si)Z(si)∑ni=1w(si)
where weights for observations are computed according to their distance to the interpolation location,
w(si)=||si−s0||−p
with ||?|| indicating Euclidean distance and p an inverse distance weighting power, defaulting to 2. If s0 coincides with an observation location, the observed value is returned to avoid infinite weights. The inverse distance power determines the degree to which the nearer point(s) are preferred over more distant points; for large values IDW converges to the one-nearest-neighbor interpolation.
The corresponding gstat function is idw(). The function
takes a formula, a location dataset (sf) and a
grid (stars). The formula objects is
consistent with the formulas we used for specifying regression models.
So, here we predict zinc (response). The ~ 1
part means that we do not include any additional predictors/covariates.
The predictions are only based on the spatial locations.
zn.idw <- gstat::idw(zinc ~ 1, locations=meuse, newdata=meuse_gridcv, idp = 2)## [inverse distance weighted interpolation]The output is a stars object with two attributes: 1)
var1.pred contains the predicted values, and 2)
var1.var contains the variance estimates. The statistics of
var1.var are NA because inverse distance does
not provide prediction error variances.
zn.idw## stars object with 2 dimensions and 2 attributes
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## var1.pred 128.4345 293.2323 371.4119 423.1647 499.8198 1805.776 5009
## var1.var NA NA NA NaN NA NA 8112
## dimension(s):
## from to offset delta refsys x/y
## x 1 78 178440 40 Double_Stereographic [x]
## y 1 104 333760 -40 Double_Stereographic [y]We can plot the predictions. Here, I first convert them with
rast() to a SpatRaster from the
terra package. Overplotting the sample locations didn’t
work right otherwise. This is a bug and should probably not be needed,
but the example shows that you can convert back and forth between
stars and terra.
plot(rast(zn.idw["var1.pred"]))
plot(meuse["zinc"], col=1, cex=0.5, add=T, type="p")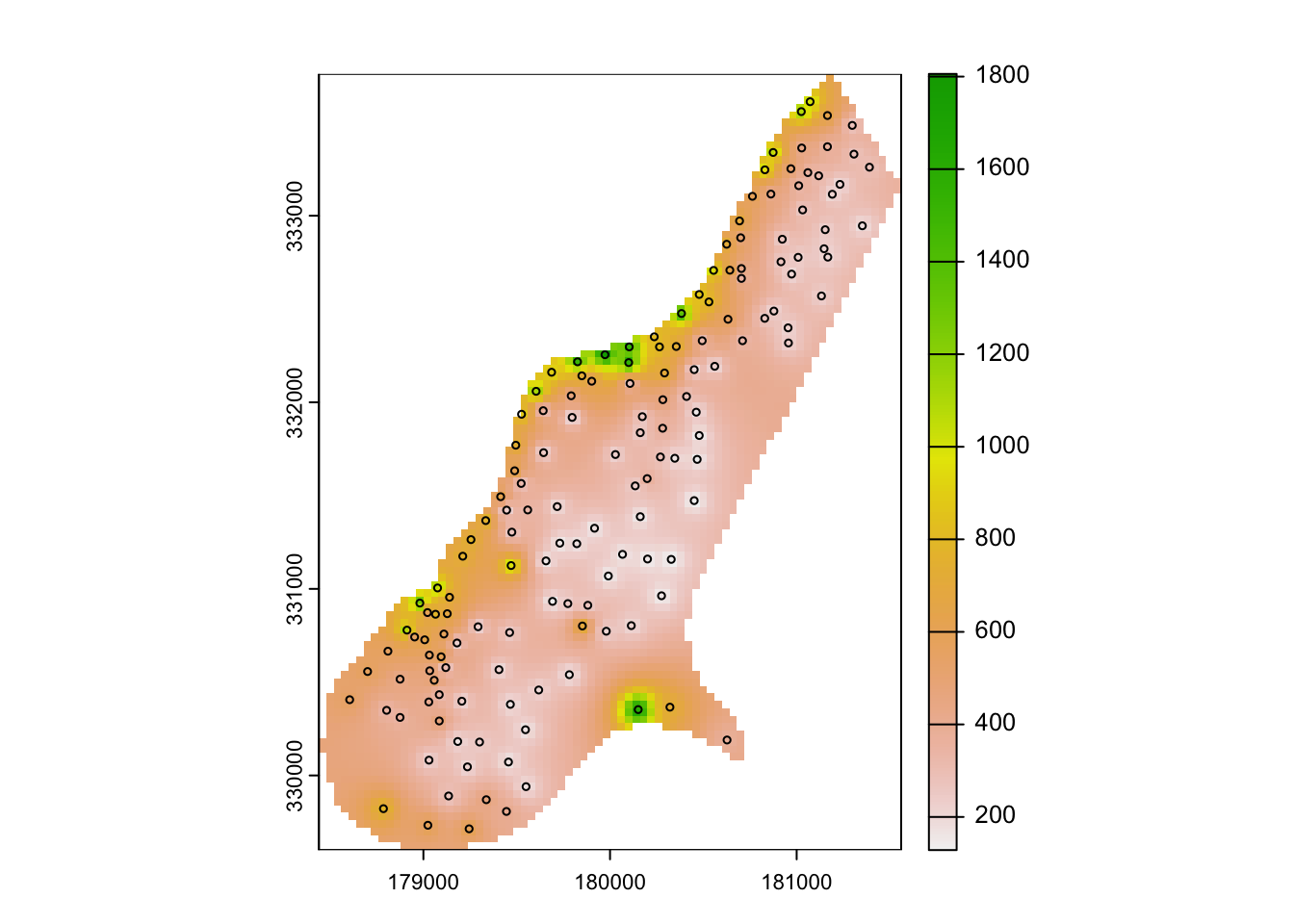
# mapview(zn.idw, zcol='var1.pred', layer.name = "Zinc ppm")IDW considers only distances to the prediction location and thus ignores the spatial configuration of observations. This may lead to undesired effects if the observation locations are strongly clustered.
Notably, IDW weights are guaranteed to be between 0 and 1, resulting in interpolated values never outside the range of observed values.
Stationarity
The spatial correlation between two observations of a variable z(s) at locations s1 and s2 cannot be estimated, as only a single pair is available. To obtain the necessary replication, we need to replicate the experiment with multiple samples from multiple locations. But to do that, we need to assume that the spatial process is the same at all those locations. In other words, we need to make stationarity assumptions about our spatial process.
Stationarity means that characteristics of a random function (our spatial process that generates a variable) stay the same when shifting a given set of n points from one part of a region to another. The spatial process is called strictly stationary if for any set of n points s1,...,sn the multivariate distribution does not change. In other words, everything looks the same everywhere and anytime. Well, this may not seem like a reasonable assumption for geographic processes.
A less restrictive condition is given by weak stationarity, which is also called second-order stationarity because it only assumes stationarity of the first two moments of the distribution. That is, the spatial process has a constant mean E[Z(x)] and variance Var[Z(x)]. Also, the covariance between two observations separated by a distance h: cov(Z(x+h),Z(x)) only relies on the distance h between the observations and not on the spatial location x inside the region.
A specific type of second-order stationarity, and most important for the variogram analysis, is called intrinsic stationarity. Here, we assume second-order stationarity of the differences between pairs of values at two locations: Z(x+h)−Z(x). We are not interested in Z(x) but in the differences. Again, this implies the assumption that the variance of these differences does not depend on location but only depends on separation distance h:
Var(Z(x+h)−Z(x))=2γ(h)
Lagged scatter plots
A simple way to acknowledge that spatial correlation is present or not is to make scatter plots of pairs Z(si) and Z(sj), grouped according to their separation distance hij=||si−sj||.
gstat::hscat(log(zinc) ~ 1, meuse, (0:9) * 100)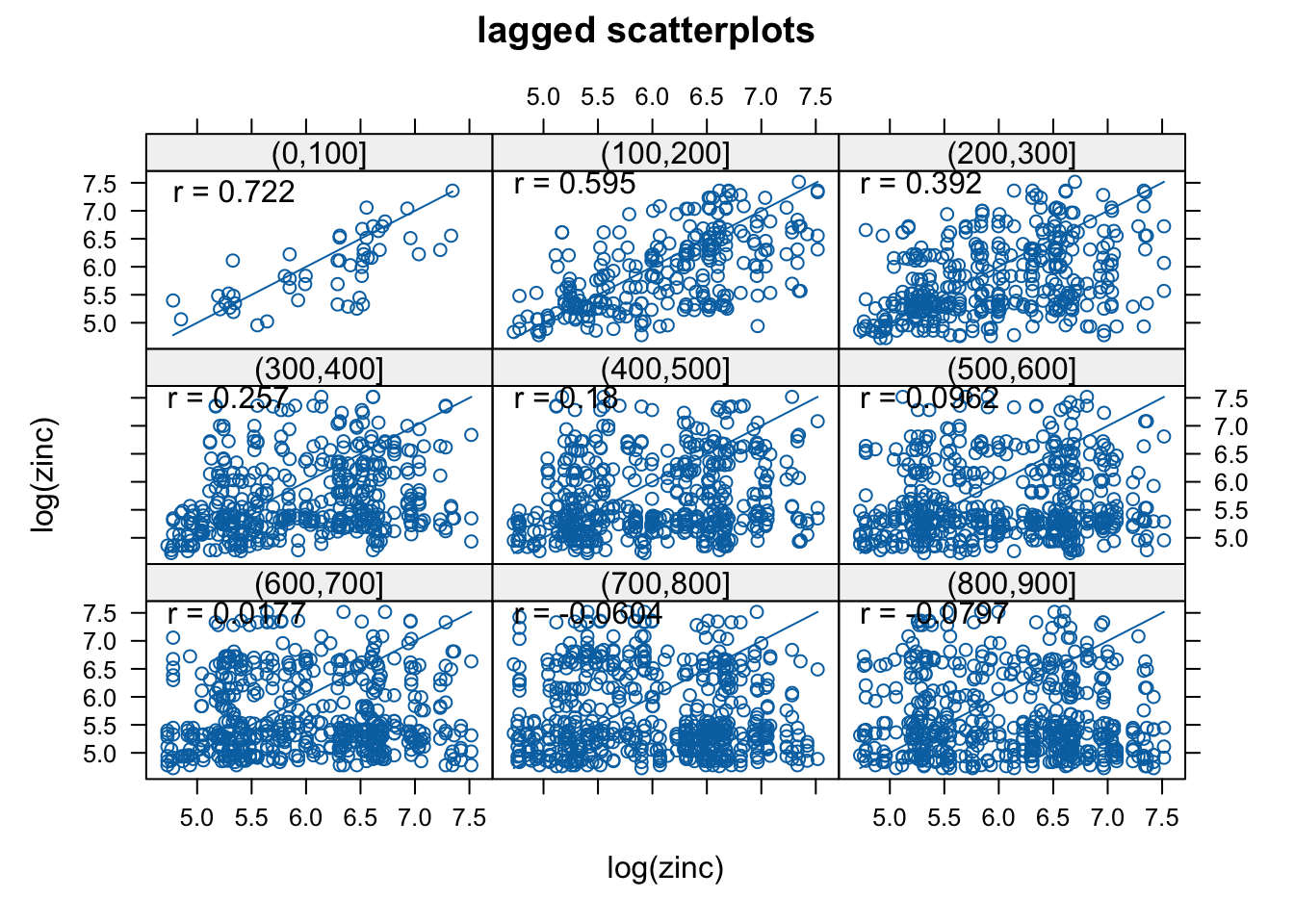
Variogram cloud
We can plot the difference between pairs of sample values as a function of separation distance h. The so-called variogram cloud plots all possible squared differences of observation pairs (Z(x)−Z(x+h))2 against their separation distance h. The difference is divided by 2 to account for the fact that two points share this value. Hence, γ is often called semi-variance (half the variance).
γh=12(Z(x)−Z(x+h))2
meuse.vc <- variogram(log(zinc) ~ 1, meuse, cloud = TRUE)
plot(meuse.vc, ylab=bquote(gamma), xlab=c("h (separation distance in m)"))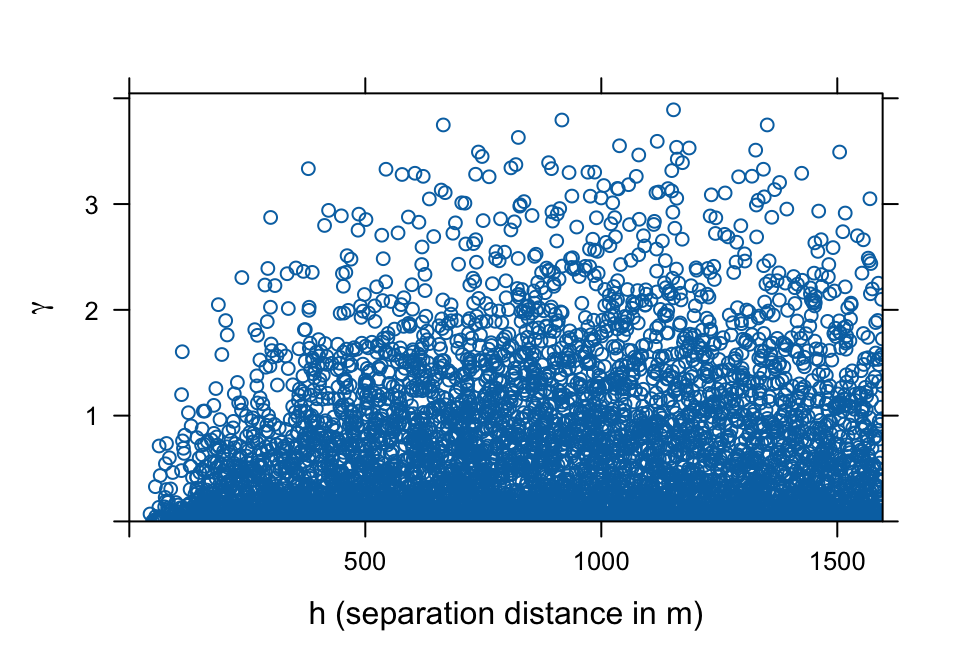
Since γ is a difference it is a measure of dissimilarity, i.e., the smaller the dissimilarity the more similar are observations. Thus, the variogram cloud allows you to observe if sample pairs closer to each other are more similar than pairs further apart, which is usually the case. The variogram cloud also allows you to observe the distribution of dissimilarity at particular distances. For example, the variogram cloud above shows that γ has a skewed distribution at any distance. That is, the majority of plots are similar even at higher distances. However, dissimilarity increases with distances up to 500 m or so.
Sample variogram
In geostatistics the spatial correlation is often modeled from the sample variogram. The sample variogram plots averages of the variogram cloud values over distance intervals h. If we further assume isotropy, which is direction independence of semivariance, we can replace the vector h with its length, ||h||.
Under this assumption, the variogram can be estimated from Nh sample data pairs z(x), z(x+h) for a number of distances (or distance intervals) ˜hj by:
ˆγ(˜hj)=12NhNh∑i=1(Z(x)−Z(x+h))2
You can construct a sample variogram() in R using the
variogram function of the gstat package. The ~ 1 defines a single
constant predictor leading to a spatially constant mean coefficient.
meuse.v <- gstat::variogram(log(zinc) ~ 1, meuse)
plot(meuse.v, ylab=bquote(gamma), xlab=c("h (separation distance in m)"))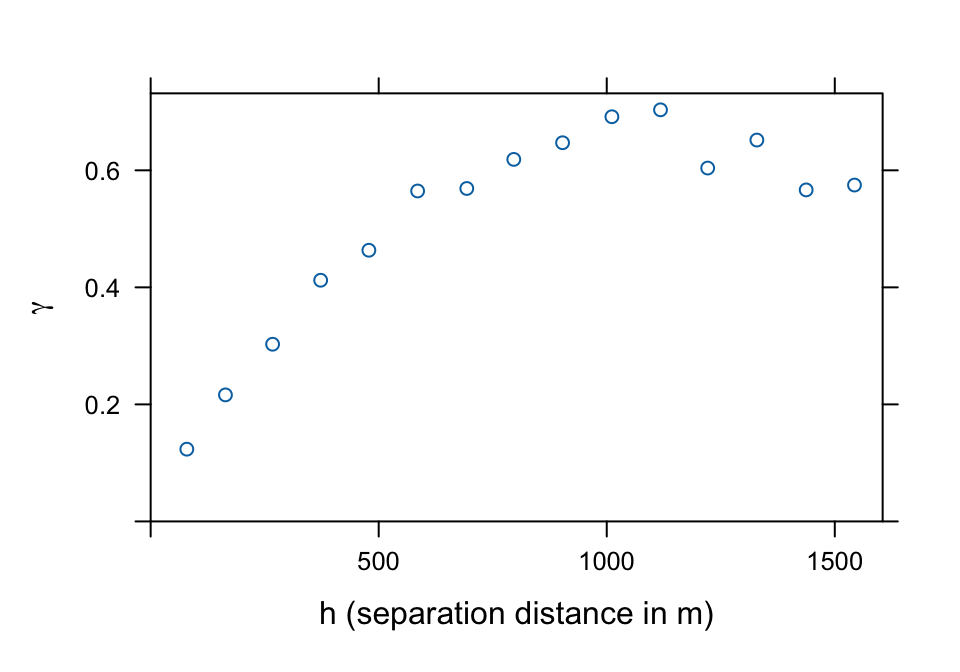
Direction dependence
The previously used variogram function makes a number of decisions by
default. It decides that direction is ignored: point pairs are merged on
the basis of distance, not direction. An alternative is, for example to
look in four different angles by specifying the alpha
keyword.
plot(variogram(log(zinc) ~ 1, meuse, alpha = c(0, 45, + 90, 135)))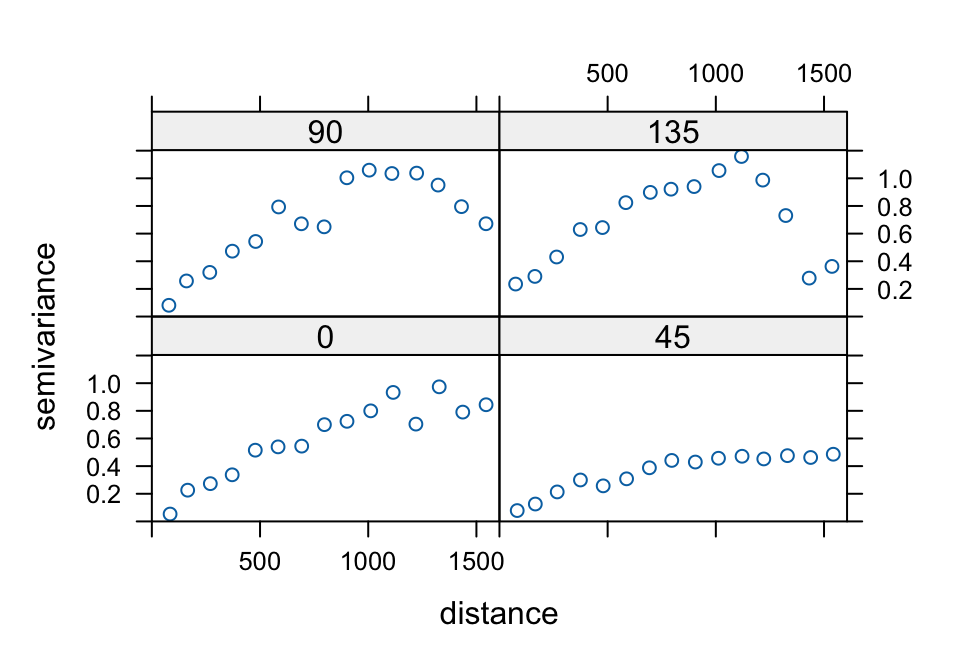
Cutoff and lag width
A similar issue is the cutoff distance, which is the maximum distance
up to which point pairs are considered and the width of distance
interval over which point pairs are averaged in bins. The default value
gstat uses for the cutoff value is one third of the largest
diagonal of the bounding box (or cube) of the data. Good reasons to
decrease the cutoff may be when a local prediction method is foreseen,
and only semivariances up to a rather small distance are required. For
the interval width, gstat uses a default of the cutoff
value divided by 15.
plot(variogram(log(zinc) ~ 1, meuse, cutoff = 1000, width = 50))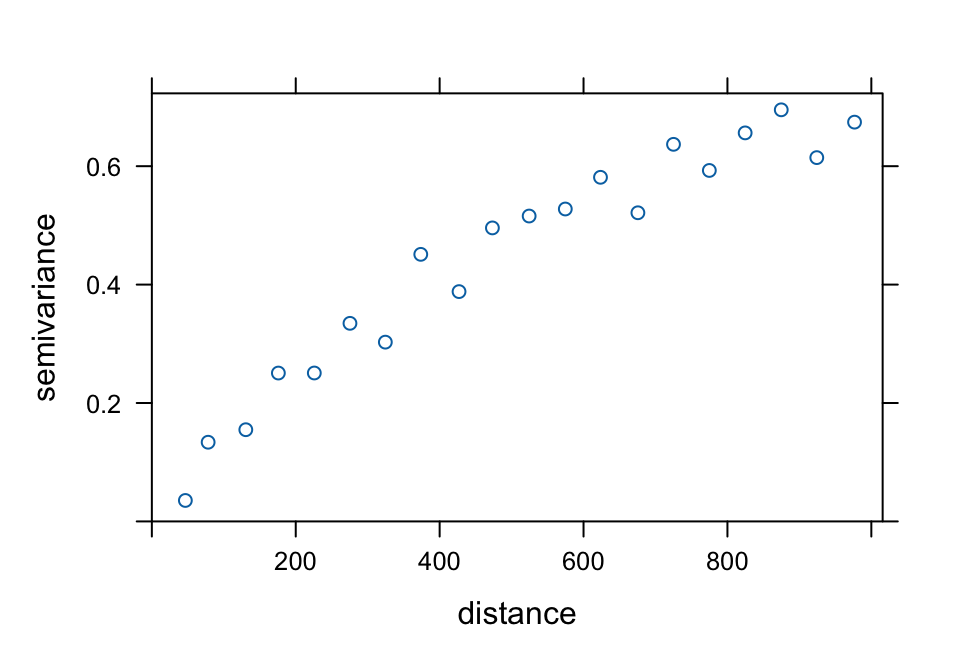
Theoretical variogram
Using the sample variogram (also called experimental variogram), we can fit a theoretical variogram model to estimate a number of parameters that describe the range of autocorrelation and variance:
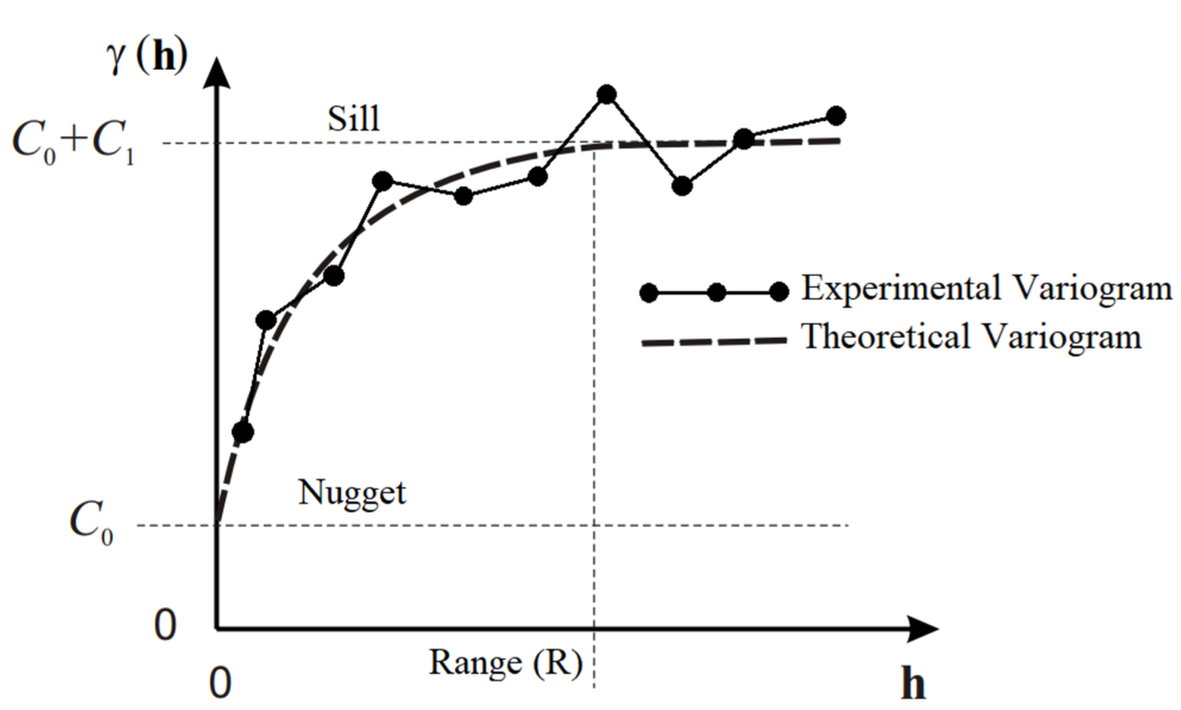
- R is the range of spatial auto-correlation, i.e., sample locations separated by distances closer than R are spatially auto-correlated, whereas locations farther apart than R are not.
- C0 is the nugget effect, which can be attributed to measurement errors or spatial sources of variation at distances smaller than the sampling interval.
- C1 is the partial sill (sill minus nugget).
- C0+C1 is the sill, which is the modeled semi-variance at range R.
A wide range of variogram models are available for estimating these
parameters, e.g., linear, spherical, exponential, Gaussian. Note, the
exponential, Gaussian, and Bessel model reach their sill asymptotically
(as h→∞). For an overview
of all available variogram models in R, type in the command line
vgm() without any model arguments.
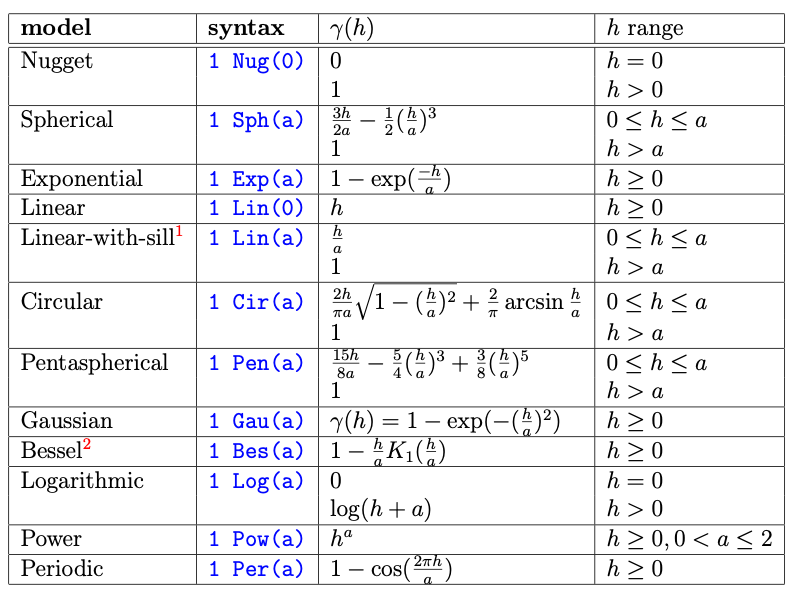
Pebesma, E. (2014): gstat user’s manual
Use the vgm() function to construct a variogram model by
specifying the nugget C0, partial
sill C1 (psill),
range R, and the model
form.
myVariogramModel <- vgm(psill=1, "Sph", range=100, nugget=0.5)
plot(myVariogramModel, cutoff=150)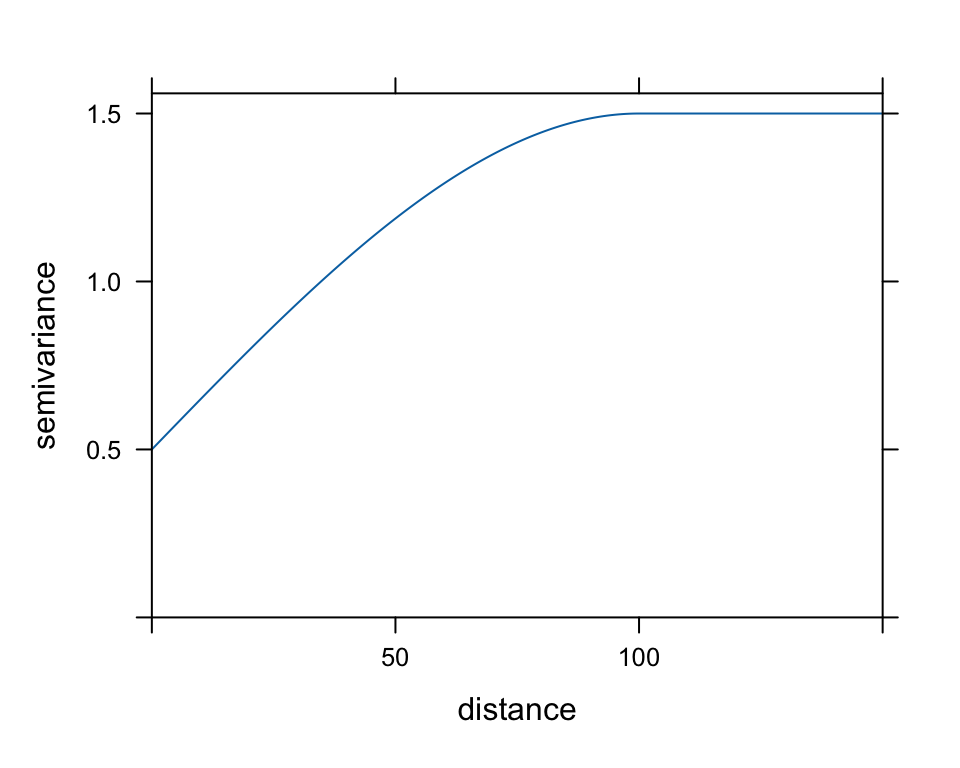
A visual overview of the basic variogram models available in
gstat can be obtained as follows:
show.vgms()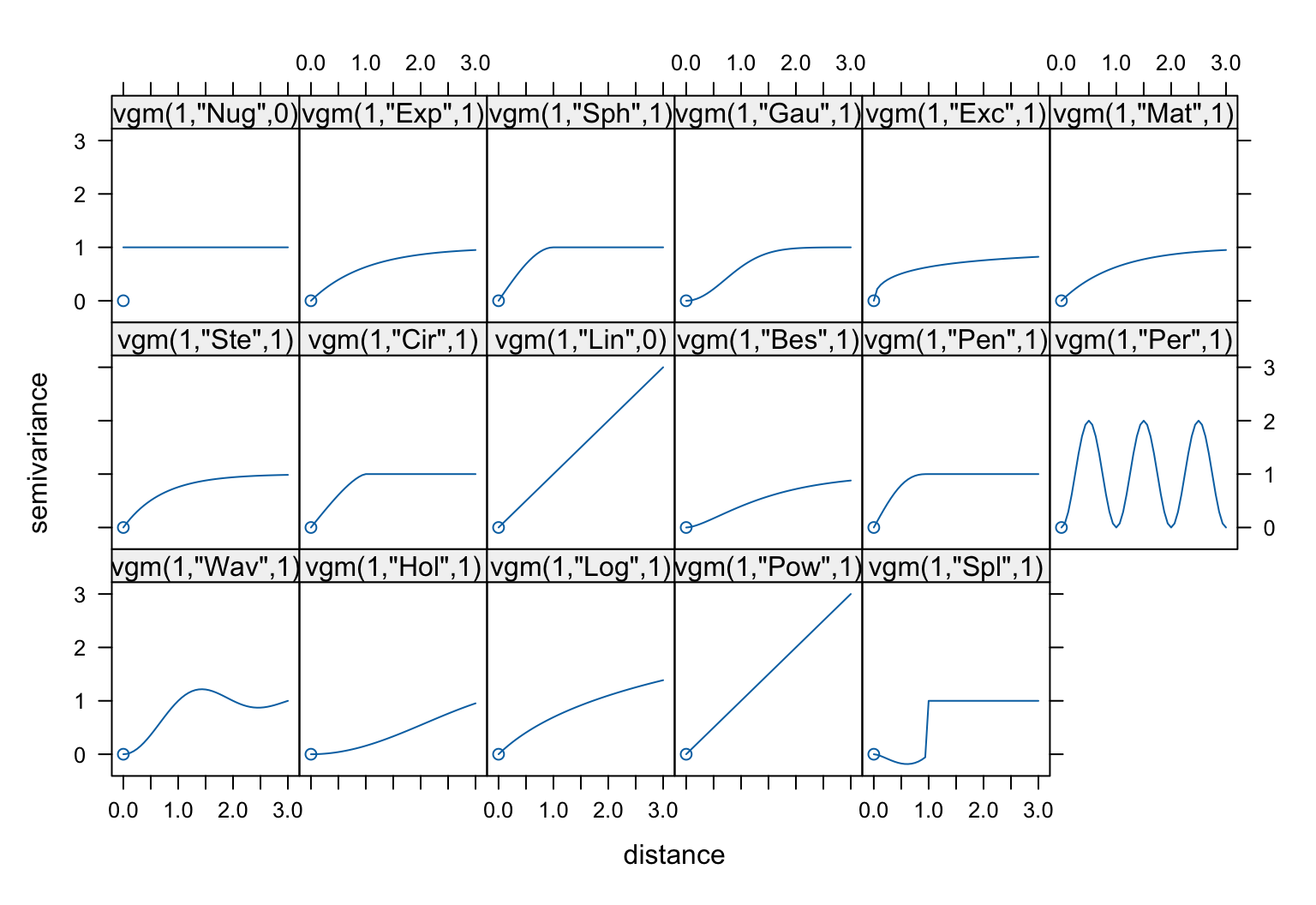
Fit a variogram
For weighted least squares fitting of a variogram model to the sample variogram, we need to take several steps: 1) calculate the sample variogram, 2) choose a suitable model, 3) choose initial parameters for our model, 4) and fit the model.
- Calculate the sample variogram
meuse.v <- variogram(log(zinc) ~ 1, meuse)Choose a suitable model (e.g. exponential, spherical), with or without nugget
Choose suitable initial values for partial sill(s), range(s), and possibly nugget
Let’s try the the spherical model with and some initial guesses for the parameters. Looking at the sample variogram, good initial values for the nugget may be 0.1, for the partial sill (sill minus nugget) = 0.6, and for the range perhaps 800m. We can plot both, the sample variogram and model on top of each other to evaluate our initial model.
meuse.sph <- vgm(psill=0.6, "Sph", range=800, nugget=0.1)
plot(meuse.v, meuse.sph, cutoff=1000)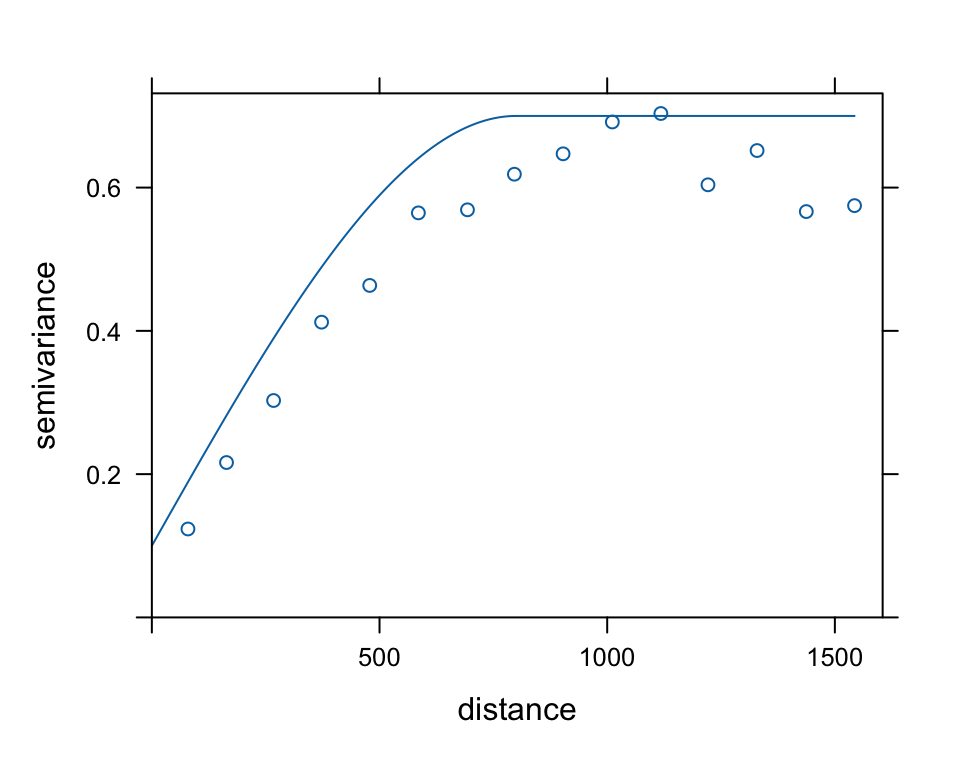
- Fit this model using one of the fitting criteria
meuse.vfit <- fit.variogram(meuse.v, meuse.sph)
meuse.vfit## model psill range
## 1 Nug 0.05065661 0.0000
## 2 Sph 0.59060200 896.9784Let’s plot the fitted variogram over the sample variogram:
plot(meuse.v, meuse.vfit)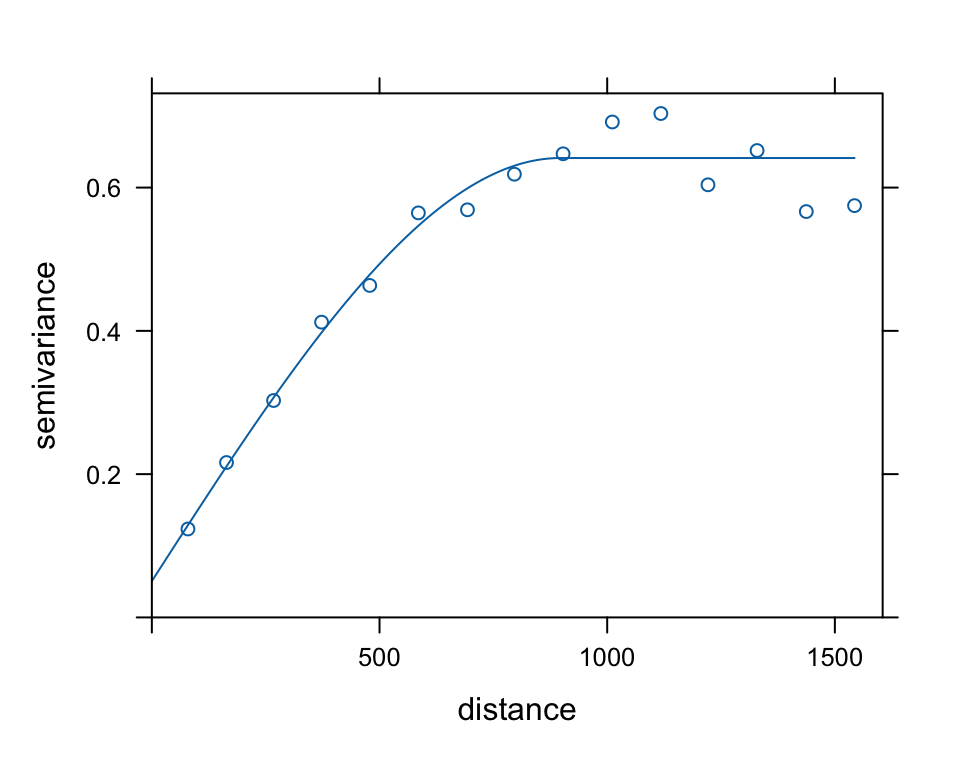
Model fitting may fail if we choose initial values too far off from reasonable values.
fit.variogram(meuse.v, vgm(1, "Sph", 10, 1))## Warning in fit.variogram(meuse.v, vgm(1, "Sph", 10, 1)): singular model in variogram fit## model psill range
## 1 Nug 1 0
## 2 Sph 1 10Partial variogram model fitting
Also partial fitting of variogram coefficients can be done with
gstat. Suppose we know for some reason that the partial
sill for the nugget model (i.e. the nugget variance) is 0.06, and we
want to fit the remaining parameters. Alternatively, the range
parameter(s) can be fixed using argument fit.ranges.
fit.variogram(meuse.v, vgm(0.6, "Sph", 800, 0.06), fit.sills = c(FALSE, TRUE))## model psill range
## 1 Nug 0.0600000 0.0000
## 2 Sph 0.5845857 923.0126Visual variogram model fitting
An alternative approach to fitting variograms is by visual fitting,
the so-called eyeball fit. Package geoR provides a
graphical user interface for inter-actively adjusting the
parameters:
library(geoR)
library(sp)
meuse_sp <- as(meuse["zinc"], "Spatial")
v.eye <- eyefit(variog(as.geodata(meuse_sp), max.dist = 1500))
ve.fit <- as.vgm.variomodel(v.eye[[1]])Typically, visual fitting will minimize |γ(h)−ˆγ(h)| with emphasis on short distance/small γ(h) values, as opposed to a weighted squared difference, used by most numerical fitting. An argument to prefer visual fitting over numerical fitting may be that the person who fits has knowledge that goes beyond the information in the data.
Ordinary kriging
Kriging utilizes the theoretical variogram to interpolate values at
any location based on distant-variance relationship. We’ll perform
Ordinary Kriging at the meuse grid locations. Recall, ordinary kriging
has a constant intercept, denoted in the formula with
~ 1:
# create sample variogram
meuse.v <- gstat::variogram(log(zinc) ~ 1, meuse)
# fit variogram model
meuse.vfit <- gstat::fit.variogram(meuse.v, vgm(1, "Sph", 800, 1))
# ordinary kriging
lz.ok <- gstat::krige(log(zinc) ~ 1, meuse, meuse_gridcv, meuse.vfit)## [using ordinary kriging]plot(rast(lz.ok['var1.pred']))
plot(meuse["zinc"], col="blue", cex=0.5, type="p", add=T)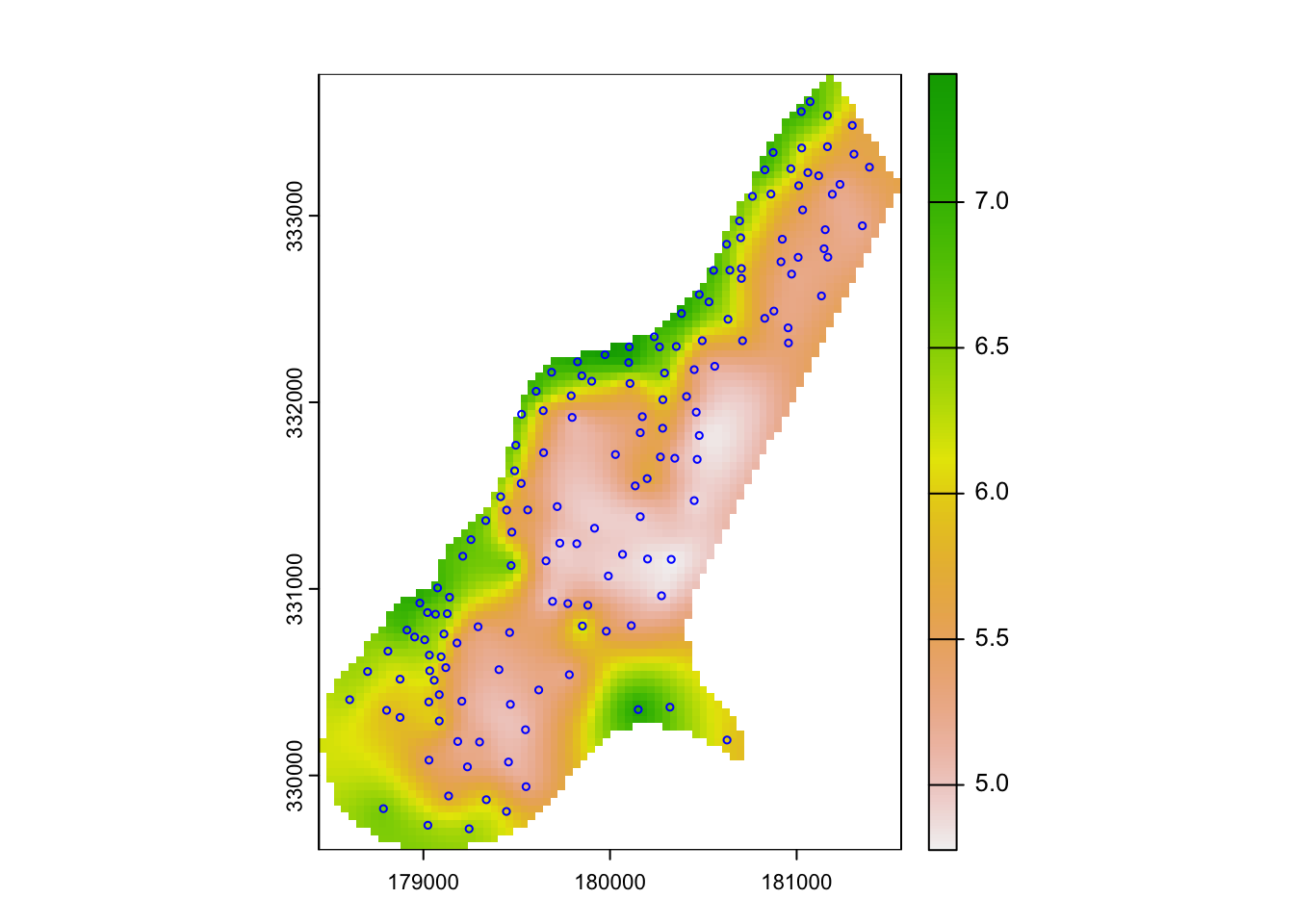
# mapview(lz.ok, zcol='var1.pred') # + mapview(meuse, zcol='zinc')Kriged maps using a variogram with no or a small nugget are typically very similar to inverse distance interpolation results.
The lz.ok object stores not just the interpolated values, but the variance values as well. These can be passed to the raster object for mapping as follows:
plot(rast(lz.ok['var1.var']))
plot(meuse["zinc"], col="blue", cex=0.5, type="p", add=T)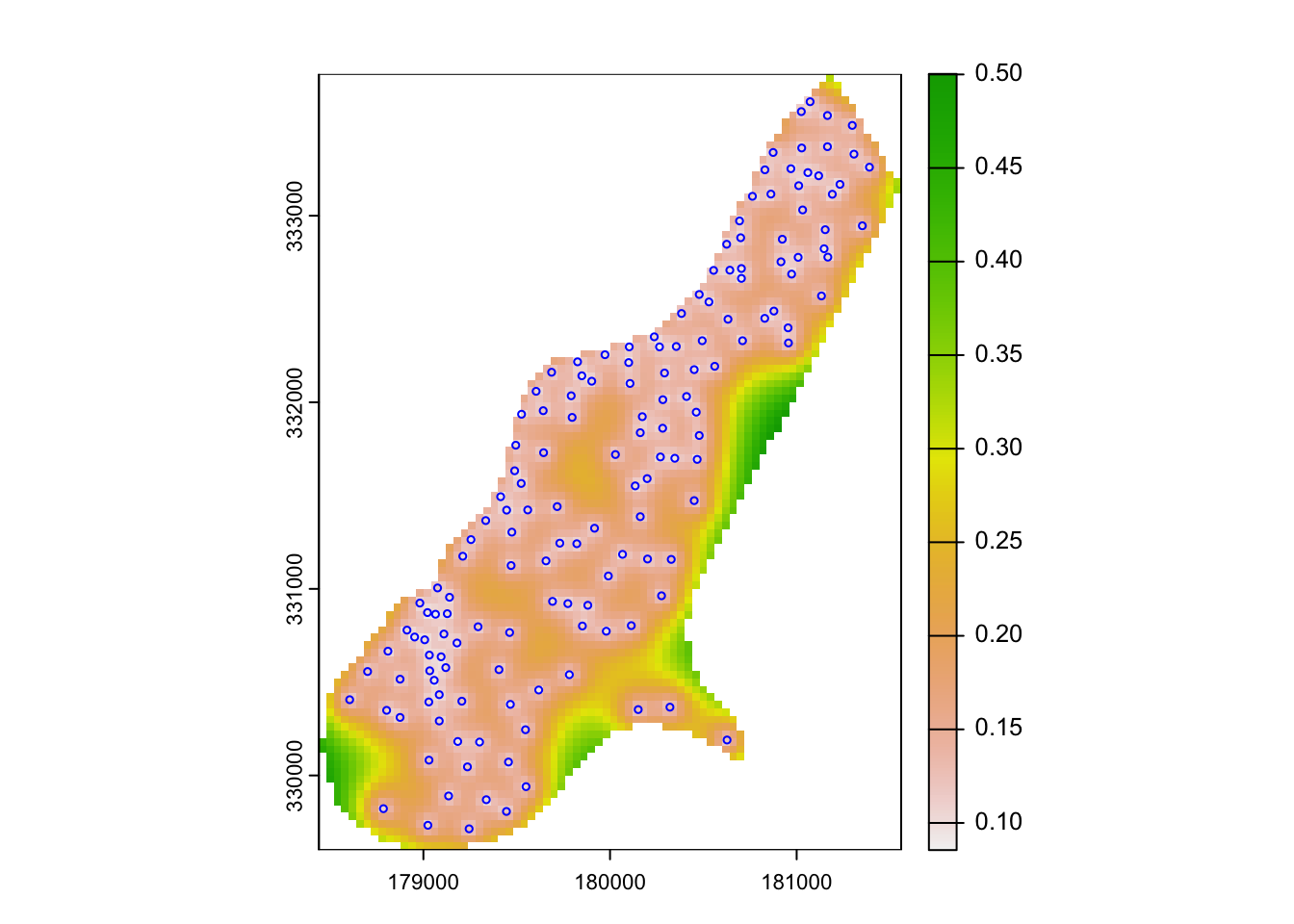
Universal kriging
Universal kriging includes other covariates to account for potential trends, e.g. here with distance to the river meuse.
# create sample variogram
meuse.rv <- variogram(log(zinc) ~ sqrt(dist), meuse)
# fit variogram model
meuse.rvfit <- fit.variogram(meuse.rv, vgm(1, "Sph", 300, 1))To apply the model spatially, we also need a grid of those
covariates. Recall meuse_gridcv
is a raster of different covariates over the study area including a
band/attribute of distance to the river (dist).
plot(meuse_gridcv["dist"])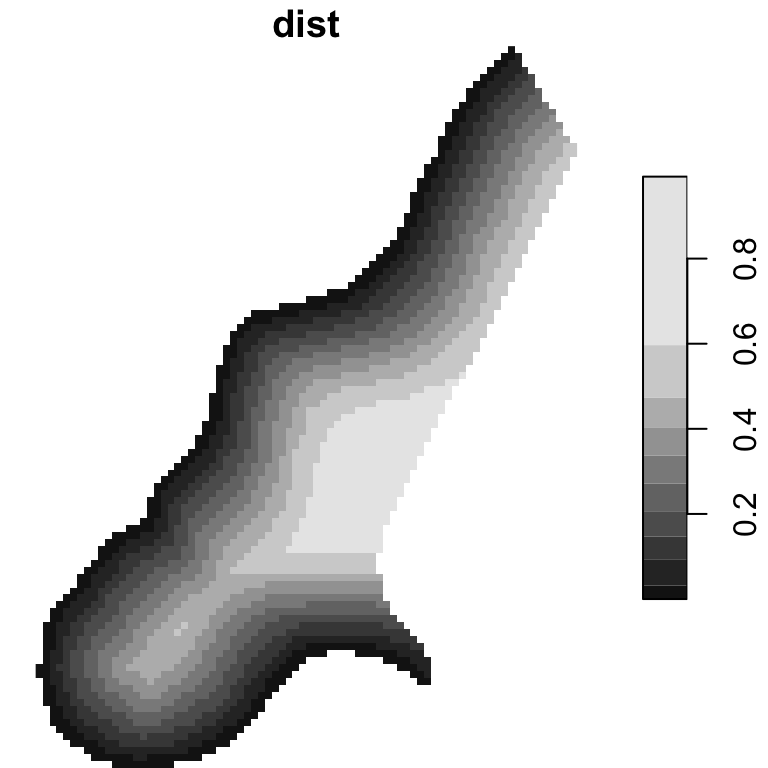
Let’s apply the model for universal kriging using the variogram model and the distance raster:
lz.uk <- krige(log(zinc) ~ sqrt(dist), locations=meuse,
newdata=meuse_gridcv,
model=meuse.rvfit)## [using universal kriging]..and plot the results.
plot(rast(lz.uk["var1.pred"]))
plot(meuse["zinc"], col="red", cex=0.5, type="p", add=T)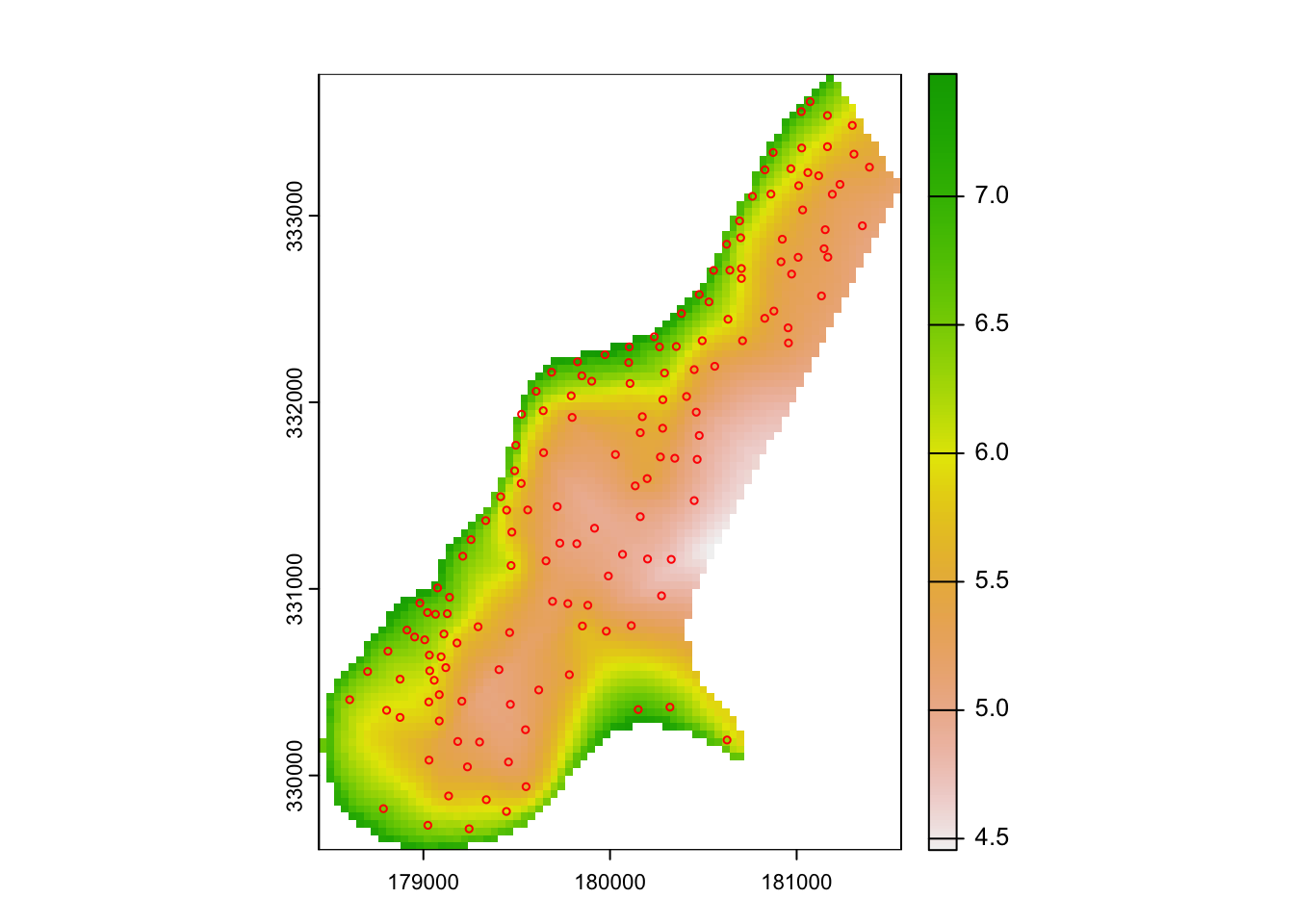
Comparison
Comparison of IDW and Kriging results. The graph shows from left to right, IDW, Ordinary Kriging and Universal Kriging results.
# simple back-transformation
lz.uk_pred <- exp(rast(lz.uk["var1.pred"]))
lz.ok_pred <- exp(rast(lz.ok["var1.pred"]))
# stack all three rasters
pred_stack <- c(rast(zn.idw["var1.pred"]),
lz.ok_pred,
lz.uk_pred)
names(pred_stack) <- c("IDW", "OK", "UK")
plot(pred_stack, nc=3, range = c(0, 2000), axes=F)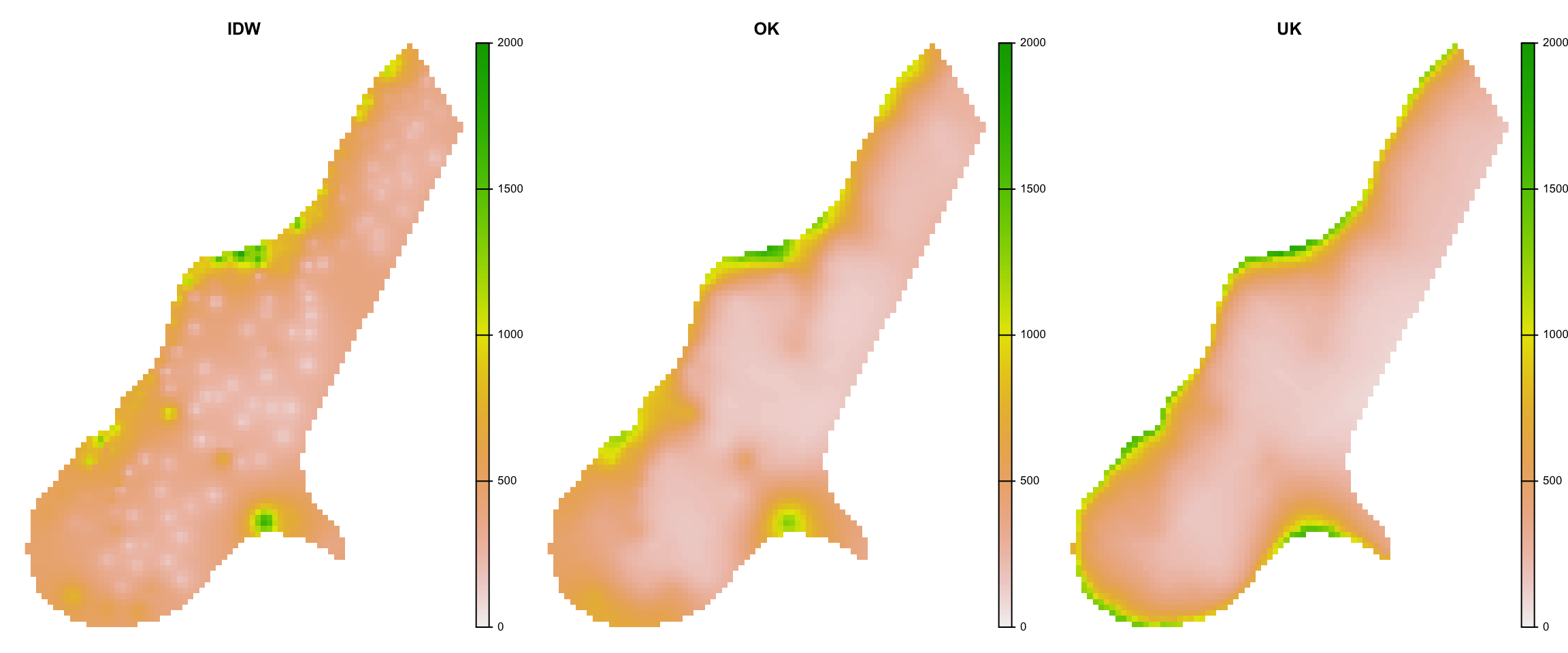
References
Bivand, R. S., Pebesma, E., Gómez-Rubio, V. (2013): Applied Spatial Data Analysis with R. Second Edition. Springer
Pebesma, E. (2022): The meuse data set: a brief tutorial for the gstat R package.
Pebesma, E. (2022): stars: Spatiotemporal Arrays, Raster and Vector Data Cubes.
Copyright © 2024 Humboldt-Universität zu Berlin. Department of Geography.