Content
- Multiple regression in R
- Understanding interactions
- Model-selection
effects packagelibrary(MuMIn) # exhaustive model selection
library(ggplot2) # data visualization
library(effects) # extract model effectsLet’s import the airquality dataset that we used in previous sessions.
# load airquality dataset
airq <- read.table("data/airquality.txt", sep = ",", dec = ".", header = TRUE)
# remove observations with missing data
airq <- na.omit(airq)
# log-transform Ozone
airq$logOzone <- log(airq$Ozone)
# convert the numeric Month column into a factor variable
airq$Month <- factor(airq$Month, levels = 5:9,
labels = c("May", "Jun", "Jul", "Aug", "Sep"))
# print first 5 rows of the dataset on screen
head(airq)## ID Ozone Solar Wind Temp Month Day logOzone
## 1 1 41 190 7.4 67 May 1 3.713572
## 2 2 36 118 8.0 72 May 2 3.583519
## 3 3 12 149 12.6 74 May 3 2.484907
## 4 4 18 313 11.5 62 May 4 2.890372
## 7 7 23 299 8.6 65 May 7 3.135494
## 8 8 19 99 13.8 59 May 8 2.944439Before constructing potentially complex linear models, it is important to visually explore your data and ask questions like:
What is the relationship between my response and predictor variables. Do I need to transform variables?
What is the relationship among my predictor variables? Do I need all? Do I need to account for collinearity?
You can evaluate the correlation between all variables by applying
the cor() function to a data frame. With the methods
argument, you can chose between the “pearson” (default), “kendall” and
“spearman” correlation coefficients. Pearson measures linear
correlation, while Kendall and Spearmann measure rank correlation and
hence will pick up non-linear correlation.
cor(airq[, 1:4])## ID Ozone Solar Wind
## ID 1.00000000 0.1375667 -0.08457572 -0.1803616
## Ozone 0.13756674 1.0000000 0.34834169 -0.6124966
## Solar -0.08457572 0.3483417 1.00000000 -0.1271835
## Wind -0.18036162 -0.6124966 -0.12718345 1.0000000You can obtain pairwise scatterplots between all the variables in the
data set using the function pairs(). Note, package
GGally provides the more sophisticated
ggpairs() function.
pairs(airq[,1:4])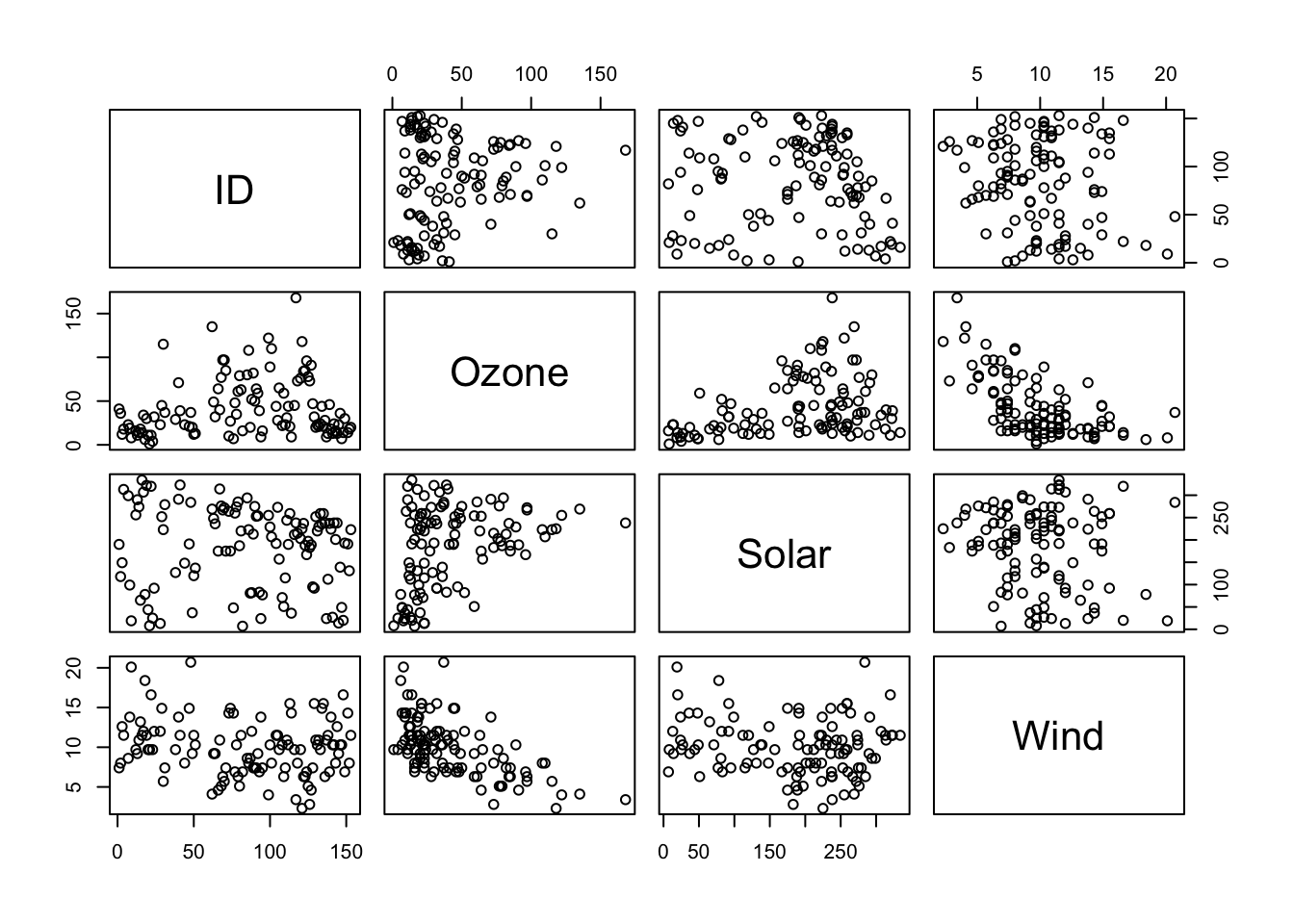
Specification of a multiple regression analysis is done by setting up
a model formula with + between the explanatory variables.
The + means that we are constructing a model where the
predictors have an additive effect on the response
variable. Note, the intercept is explicitly included in this formula. We
could include it explicitly by writing
logOzone ~ 1 + Temp + Solar + Wind. If for some reason we
wish to omit an intercept then we write
logOzone ~ 0 + Temp + Solar + Wind.
lm1 <- lm(logOzone ~ Temp + Solar + Wind, data = airq)
lm1##
## Call:
## lm(formula = logOzone ~ Temp + Solar + Wind, data = airq)
##
## Coefficients:
## (Intercept) Temp Solar Wind
## -0.262132 0.049171 0.002515 -0.061562We can add more complex terms, i.e. polynomials, “on-the-fly” using
the I() function. The I() function “protects”
the calculation inside the I() function call, such that
operators like +, -, * and
^ are not misinterpreted as formula operators.
lm2 <- lm(logOzone ~ Temp + Solar + I(Solar^2) + Wind, data = airq)
lm2##
## Call:
## lm(formula = logOzone ~ Temp + Solar + I(Solar^2) + Wind, data = airq)
##
## Coefficients:
## (Intercept) Temp Solar I(Solar^2) Wind
## -2.369e-01 4.492e-02 7.259e-03 -1.413e-05 -5.876e-02You can use the update() function to add or remove
predictors from a model:
# add a term from model lm2
lm3 <- update(lm2, . ~ . + I(Temp^2))
lm3##
## Call:
## lm(formula = logOzone ~ Temp + Solar + I(Solar^2) + Wind + I(Temp^2),
## data = airq)
##
## Coefficients:
## (Intercept) Temp Solar I(Solar^2) Wind I(Temp^2)
## 3.932e+00 -6.687e-02 7.649e-03 -1.508e-05 -5.635e-02 7.280e-04# remove a term from model lm3
lm4 <- update(lm3, . ~ . - Wind)
lm4##
## Call:
## lm(formula = logOzone ~ Temp + Solar + I(Solar^2) + I(Temp^2),
## data = airq)
##
## Coefficients:
## (Intercept) Temp Solar I(Solar^2) I(Temp^2)
## 3.561e+00 -8.328e-02 8.439e-03 -1.751e-05 8.983e-04A mathematical model can only work with numbers. So, how does R deal with categorical variables? Categorical predictor variables are internally recoded into multiple binary dummy variables, where each dummy variable switches a single factor level on (1) or off (0). For example, the variable Month in the airquality dataset has five factor levels: May, Jun, Jul, Aug, Sep. To illustrate the point, let’s recode the factor variable manually. Note, there are different names for dummy variables. Indicator variables is another common term.
airq$dummyJun <- ifelse(airq$Month=="Jun", 1, 0)
airq$dummyJul <- ifelse(airq$Month=="Jul", 1, 0)
airq$dummyAug <- ifelse(airq$Month=="Aug", 1, 0)
airq$dummySep <- ifelse(airq$Month=="Sep", 1, 0)
head(airq[!duplicated(airq$Month), -1])## Ozone Solar Wind Temp Month Day logOzone dummyJun dummyJul dummyAug dummySep
## 1 41 190 7.4 67 May 1 3.713572 0 0 0 0
## 38 29 127 9.7 82 Jun 7 3.367296 1 0 0 0
## 62 135 269 4.1 84 Jul 1 4.905275 0 1 0 0
## 93 39 83 6.9 81 Aug 1 3.663562 0 0 1 0
## 124 96 167 6.9 91 Sep 1 4.564348 0 0 0 1We do not need to create a dummy for month May since month May is
automatically coded when all other dummy variables are 0. So, if a
variable has n factors, you need n-1 dummy
variables. That also means the more factor levels the more parameters
your model needs to estimate. Let’s build a linear regression model
between logOzone and Temperature and Month using the dummy
variables.
lm4 <- lm(logOzone ~ Temp + dummyJun + dummyJul + dummyAug + dummySep, data = airq)
summary(lm4)##
## Call:
## lm(formula = logOzone ~ Temp + dummyJun + dummyJul + dummyAug +
## dummySep, data = airq)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.26925 -0.29321 0.03879 0.34206 1.50462
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.138938 0.546860 -3.911 0.000163 ***
## Temp 0.074715 0.008037 9.296 2.32e-15 ***
## dummyJun -0.468759 0.243768 -1.923 0.057192 .
## dummyJul -0.244664 0.214692 -1.140 0.257045
## dummyAug -0.293629 0.217549 -1.350 0.180009
## dummySep -0.387591 0.179445 -2.160 0.033053 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.5748 on 105 degrees of freedom
## Multiple R-squared: 0.5793, Adjusted R-squared: 0.5593
## F-statistic: 28.92 on 5 and 105 DF, p-value: < 2.2e-16Now, as mentioned before, the dummy coding is actually done internally when you include a factor variable as predictor. You do not need to do the coding manually. So, before we discuss what the output means, let’s check out the ‘automated’ code version without manual dummy coding (recall, variable Month is a factor variable):
lm5 <- lm(logOzone ~ Temp + Month, data = airq)
summary(lm5)##
## Call:
## lm(formula = logOzone ~ Temp + Month, data = airq)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.26925 -0.29321 0.03879 0.34206 1.50462
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.138938 0.546860 -3.911 0.000163 ***
## Temp 0.074715 0.008037 9.296 2.32e-15 ***
## MonthJun -0.468759 0.243768 -1.923 0.057192 .
## MonthJul -0.244664 0.214692 -1.140 0.257045
## MonthAug -0.293629 0.217549 -1.350 0.180009
## MonthSep -0.387591 0.179445 -2.160 0.033053 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.5748 on 105 degrees of freedom
## Multiple R-squared: 0.5793, Adjusted R-squared: 0.5593
## F-statistic: 28.92 on 5 and 105 DF, p-value: < 2.2e-16The model results are the same as using manually coded dummy
variables. You see the factor Month was recoded into dummy variables,
though they are now named MonthJun, MonthJul,
MonthAug, MonthSep. The first level
(May) is automatically selected as the base level (e.g. if
all other dummy variables are 0).
With the previous model, we fitted a linear regression model using
Temperature and Month as predictors. In other words, we let the linear
relationship between Temperature and logOzone vary with Month. More
precisely, we let only the intercept vary with Month. To illustrate how
we can estimate the intercepts associated with the different levels of
Month, we need to go back to our linear model equation:
logOzone=β0+β1Temp+β2MonthJun+β3MonthJul+β4MonthAug+β5Sep+ϵ
Since the dummy variables are binary, i.e. either 0 or 1, the equation above reduces to this equation for month May:
logOzone=β0+β1Temp+β2∗0+β3∗0+β4∗0+β5∗0
logOzone=β0+β1Temp
… or for Month July:
logOzone=β0+β1Temp+β2∗0+β3∗1+β4∗0+β5∗0
logOzone=β0+β1Temp+β3
Since β0 and β3 are constants, we can rearrange as follows:
logOzone=(β0+β3)+β1Temp
To estimate the different intercepts, we replace the β′s with the corresponding
parameter outputs from summary(lm5), e.g. β0 is (Intercept),
β2 is MonthJun,
β3 is MonthJul,
etc. For May the intercept of the regression line is β0 = -2.14 and for month July the
intercept is β0+β3 =
-2.14 + -0.24 = -2.38 (for month August β0+β4, etc.).
coef(lm5) # access model coefficients (beta's)## (Intercept) Temp MonthJun MonthJul MonthAug MonthSep
## -2.13893784 0.07471496 -0.46875932 -0.24466375 -0.29362919 -0.38759055lm5_intercept_May <- coef(lm5)['(Intercept)']
lm5_intercept_May## (Intercept)
## -2.138938lm5_intercept_Jul <- coef(lm5)['(Intercept)'] + coef(lm5)['MonthJul']
lm5_intercept_Jul## (Intercept)
## -2.383602lm5_slope <- coef(lm5)['Temp']
lm5_slope## Temp
## 0.07471496You see that the only parameter that changes with Month in our model is the intercept and we can calculate the intercepts based on β0 and the β’s from our dummy variables. Let’s plot the regression lines for May and July based on the calculated intercepts.
ggplot(airq, aes(x = Temp, y = logOzone)) +
geom_point(aes(col = Month)) +
scale_color_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00")) +
geom_abline(slope=lm5_slope, intercept=lm5_intercept_May, col="#E41A1C") +
geom_abline(slope=lm5_slope, intercept=lm5_intercept_Jul, col="#4DAF4A")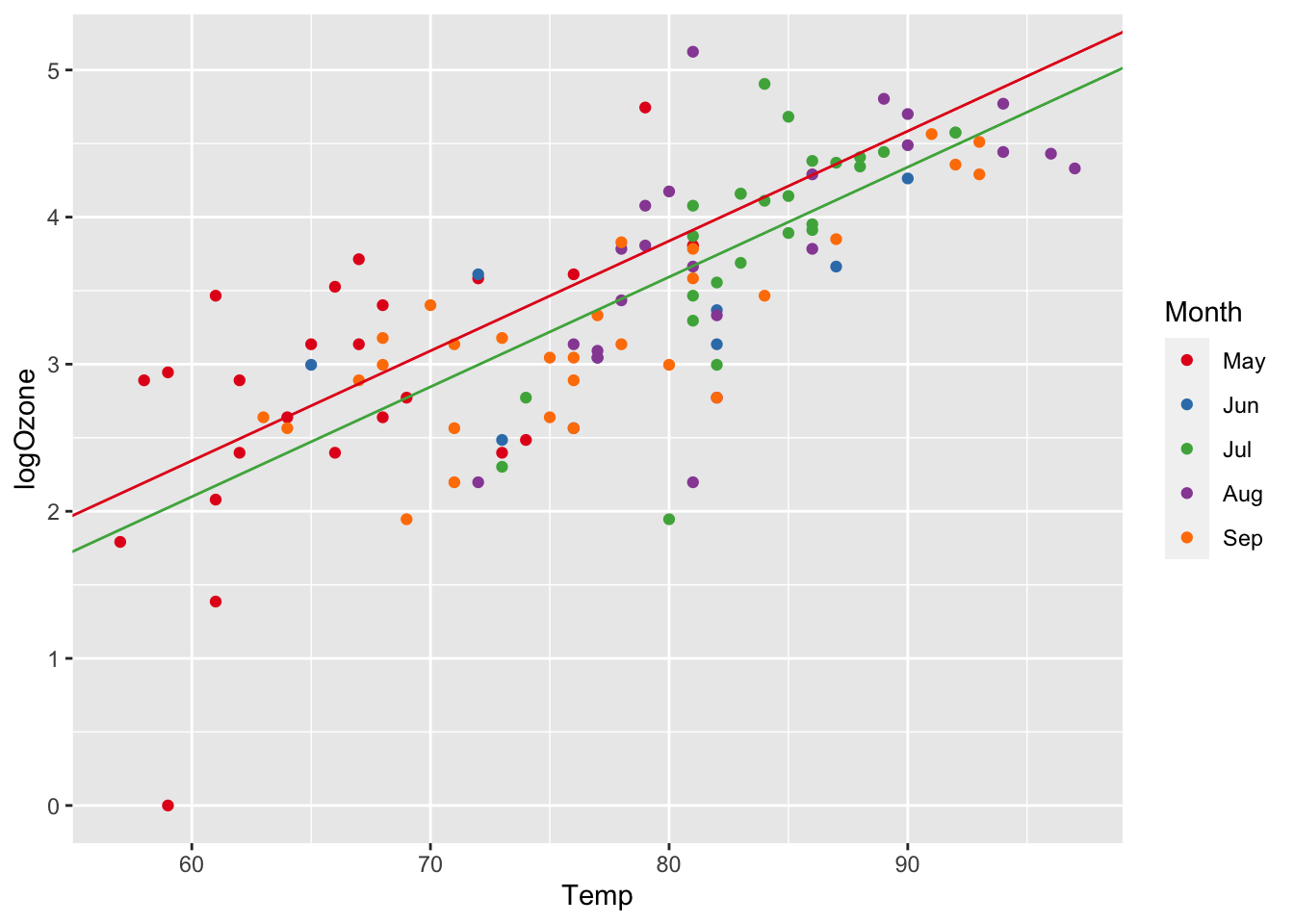
So far, we only included an additive effect of Month,
i.e. we allowed only the intercept to vary with Month. However, what if
the effect is not additive? In other words, what if the effect of Month
on Ozone is not constant? Perhaps, the effect of Month varies also with
Temperature (or the other way round), in which case we need to let the
slope vary with Month as well, not just the intercept. To
include such interactions between predictor variables, we can include
interaction terms.
In model formulas in R, interaction terms are generated using the
colon operator: var1:var2.
Usually, the interaction term is included in addition to the additive effect:
respone ~ var1+var2 + var1:var2
Rather than writing both terms, you can use the shorthand:
var1*var2. Hence, the following formula describes the same
model as the formula above:
respone ~ var1*var2
Let’s try out an interaction between Temperature and Month with our airquality dataset.
lm6 <- lm(logOzone ~ Temp * Month, data = airq)
summary(lm6)##
## Call:
## lm(formula = logOzone ~ Temp * Month, data = airq)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.19906 -0.26980 0.00246 0.35058 1.51166
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.76430 1.19238 -2.318 0.0224 *
## Temp 0.08413 0.01786 4.711 7.89e-06 ***
## MonthJun 2.42798 2.33970 1.038 0.3019
## MonthJul -3.95983 2.45826 -1.611 0.1103
## MonthAug 0.11327 1.87091 0.061 0.9518
## MonthSep 1.13404 1.54120 0.736 0.4635
## Temp:MonthJun -0.03845 0.03123 -1.231 0.2211
## Temp:MonthJul 0.04233 0.03121 1.357 0.1779
## Temp:MonthAug -0.00680 0.02477 -0.275 0.7843
## Temp:MonthSep -0.02107 0.02187 -0.963 0.3377
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.568 on 101 degrees of freedom
## Multiple R-squared: 0.6048, Adjusted R-squared: 0.5696
## F-statistic: 17.18 on 9 and 101 DF, p-value: < 2.2e-16The model output shows the parameter estimates for the base
intercept, the additive effects of Temp and Month (coded as
dummy variables), and the interaction terms with
the colon sign (:).
To estimate how the slopes vary with Month, we need to
go back to our model formula and combine the parameters as we did when
estimating the intercepts from the additive
dummy model. The parameters for the interaction terms are β6-β9.
logOzone=β0+β1∗Temp+β2∗Jun+β3∗Jul+...+β5∗Sep+β6∗Temp∗Jun+β7∗Temp∗Jul+...+β9∗Temp∗Sep
So, if we are interested in the relationship between Temperature and logOzone for May, the above equation reduces to:
logOzone=β0+β1Temp
As May is our base factor level, all other βs get multiplied by zeros and therefore eliminated. The intercept of month May is β0 and the slope is β1.
If we are interested in the relationship between Temperature and Ozone for July (or some other month), the equation reduces as follows, where β3 estimates the additive term and β6 estimates the interaction term. Note, unlike in an R formula, where the ∗ sign symbolizes the inclusion of both, additive and interaction terms, here ∗ truly represents the multiplicative sign.
logOzone=β0+β1Temp+β3Jul+β7∗Temp∗Jul
which we can rearrange as follows:
logOzone=(β0+β3)+(β1+β7)∗Temp
where the intercept is (β0+β3) and the slope is (β1+β7).
The coefficients of our airquality model with interaction term are as follows:
coef(lm6) # access model coefficients (beta's)## (Intercept) Temp MonthJun MonthJul MonthAug MonthSep Temp:MonthJun
## -2.764304030 0.084124863 2.427979206 -3.959831889 0.113272053 1.134041099 -0.038447329
## Temp:MonthJul Temp:MonthAug Temp:MonthSep
## 0.042334206 -0.006799668 -0.021065369To estimate ozone concentration for month May, we grab the intercept and slope term:
logOzoneMay=−2.76+0.08∗Temp
To estimate ozone concentration for month July, we have to add the parameters for the July intercept offset (-3.96) and the slope offset from the interaction term (0.04):
logOzoneJuly=(−2.76−3.96)+(0.08+0.04)∗Temp
We can do this directly in R and plot the corresponding regression lines:
lm6_intercept_May <- coef(lm6)['(Intercept)']
lm6_intercept_Jul <- coef(lm6)['(Intercept)'] + coef(lm6)['MonthJul']
lm6_slope_May <- coef(lm6)['Temp']
lm6_slope_Jul <- coef(lm6)['Temp'] + coef(lm6)['Temp:MonthJul']
ggplot(airq, aes(x = Temp, y = logOzone)) +
geom_point(aes(col = Month)) +
scale_color_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00")) +
geom_abline(slope=lm6_slope_May, intercept=lm6_intercept_May, col="#E41A1C") +
geom_abline(slope=lm6_slope_Jul, intercept=lm6_intercept_Jul, col="#4DAF4A")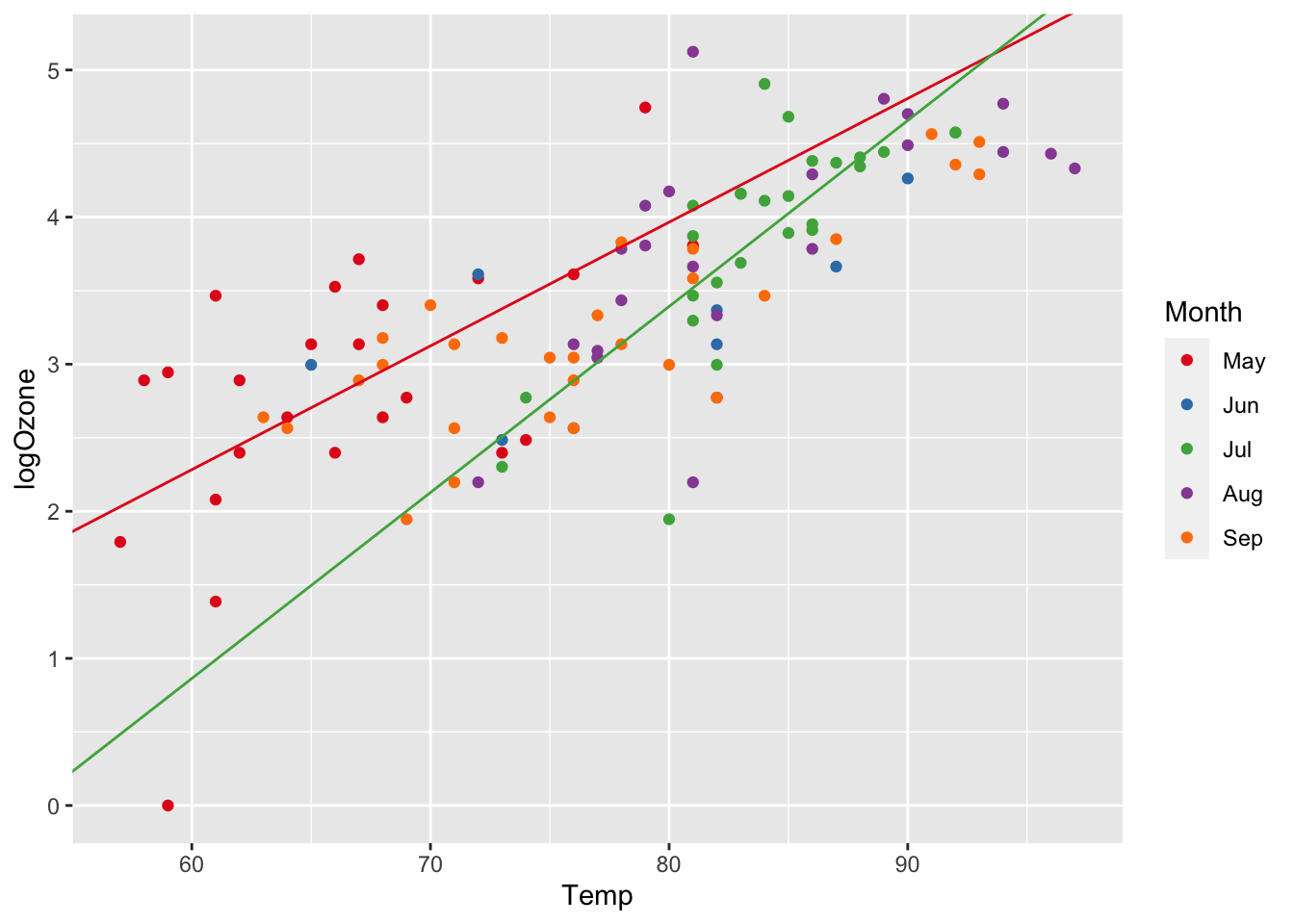
effects packageAs always, there already exists a package doing all the hard
coding-work for us. In this case, we make use of the
effect() function from the effects
package:
efct <- effects::effect(term = "Temp:Month", mod = lm6)
efct <- as.data.frame(efct) # convert list to data.frame
efct$Month <- factor(efct$Month, levels = c("May", "Jun", "Jul", "Aug", "Sep"))
head(efct)## Temp Month fit se lower upper
## 1 60 May 2.283188 0.1635384 1.958772 2.607604
## 2 70 May 3.124436 0.1320779 2.862429 3.386443
## 3 80 May 3.965685 0.2681741 3.433700 4.497670
## 4 90 May 4.806934 0.4360770 3.941874 5.671993
## 5 100 May 5.648182 0.6100672 4.437973 6.858392
## 6 60 Jun 2.404327 0.5038101 1.404903 3.403751Note: confidence intervals are calculated automatically. You can
change the level of the confidence intervals setting the argument
confint. See ?effect.
Plotting the regression on the log-scale:
ggplot(airq) +
geom_point(aes(x = Temp, y = logOzone, col = Month)) +
geom_line(data = efct, aes(x = Temp, y = fit, col = Month)) +
scale_color_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00"))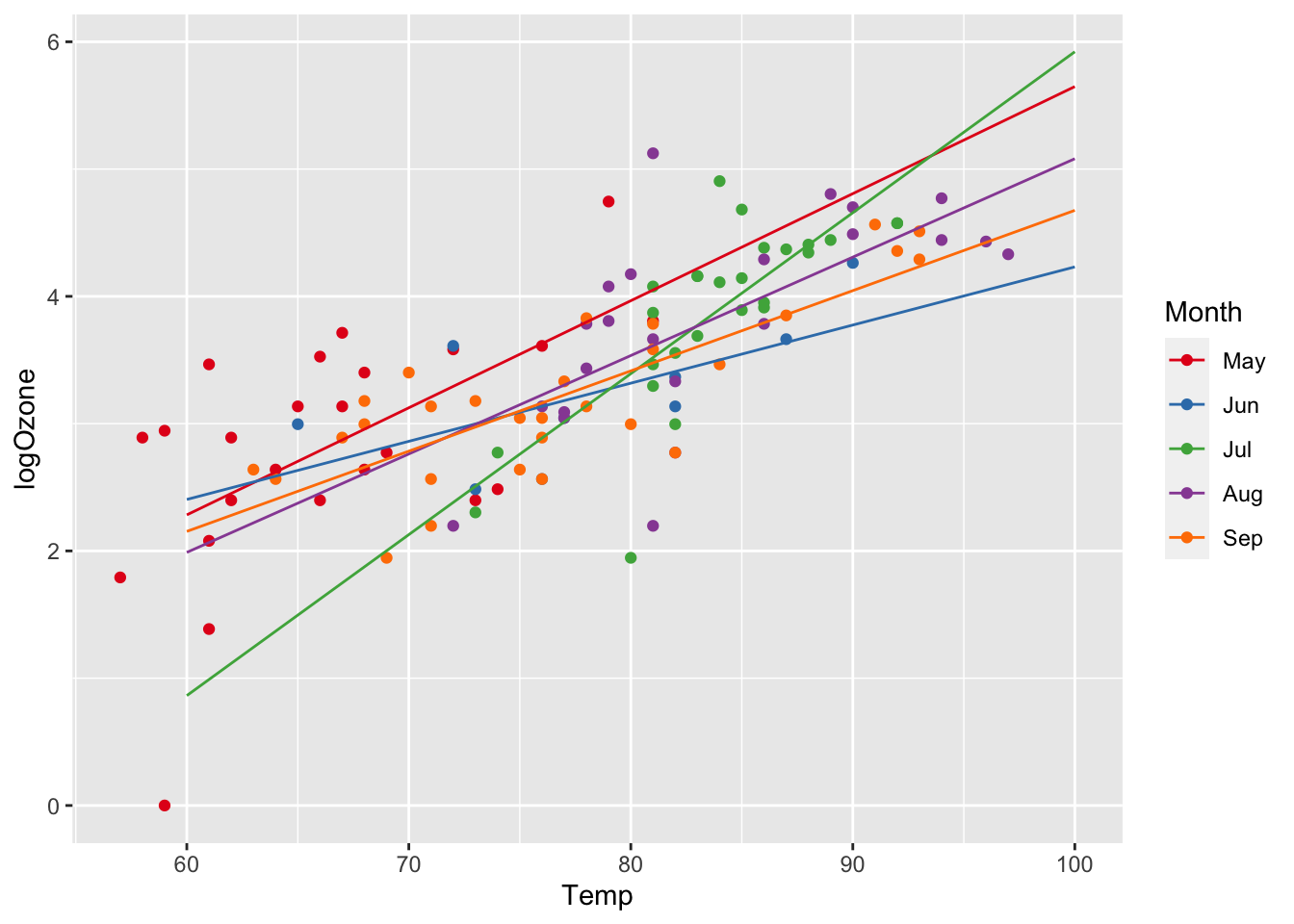
Add confidence intervals to the plot as ribbons using
geom_ribbon():
ggplot(airq) +
geom_point(aes(x = Temp, y = logOzone, col = Month)) +
geom_ribbon(data = efct, aes(x = Temp, ymin = lower, ymax = upper, fill = Month), alpha = 0.2) +
geom_line(data = efct, aes(x = Temp, y = fit, col = Month)) +
scale_color_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00")) +
scale_fill_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00"))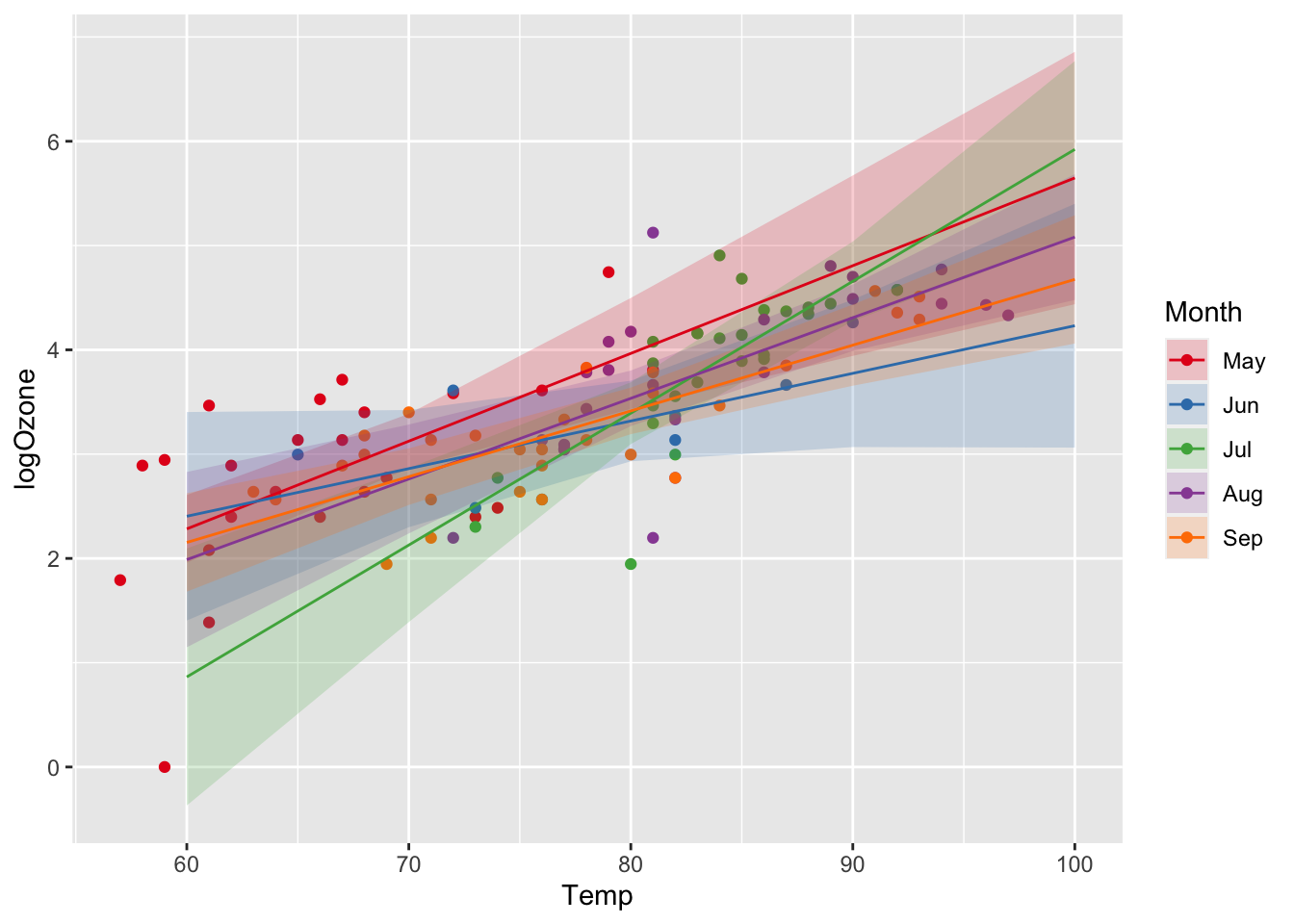
Converting predictions back to the linear scale:
efct <- effect(term = "Temp:Month", mod = lm6)
efct <- as.data.frame(efct)
efct$Month <- factor(efct$Month, levels = c("May", "Jun", "Jul", "Aug", "Sep"))
ggplot(airq, aes(x = Temp, y = Ozone, col = Month)) +
geom_point() +
scale_color_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00")) +
geom_line(data = efct, aes(y = exp(fit)))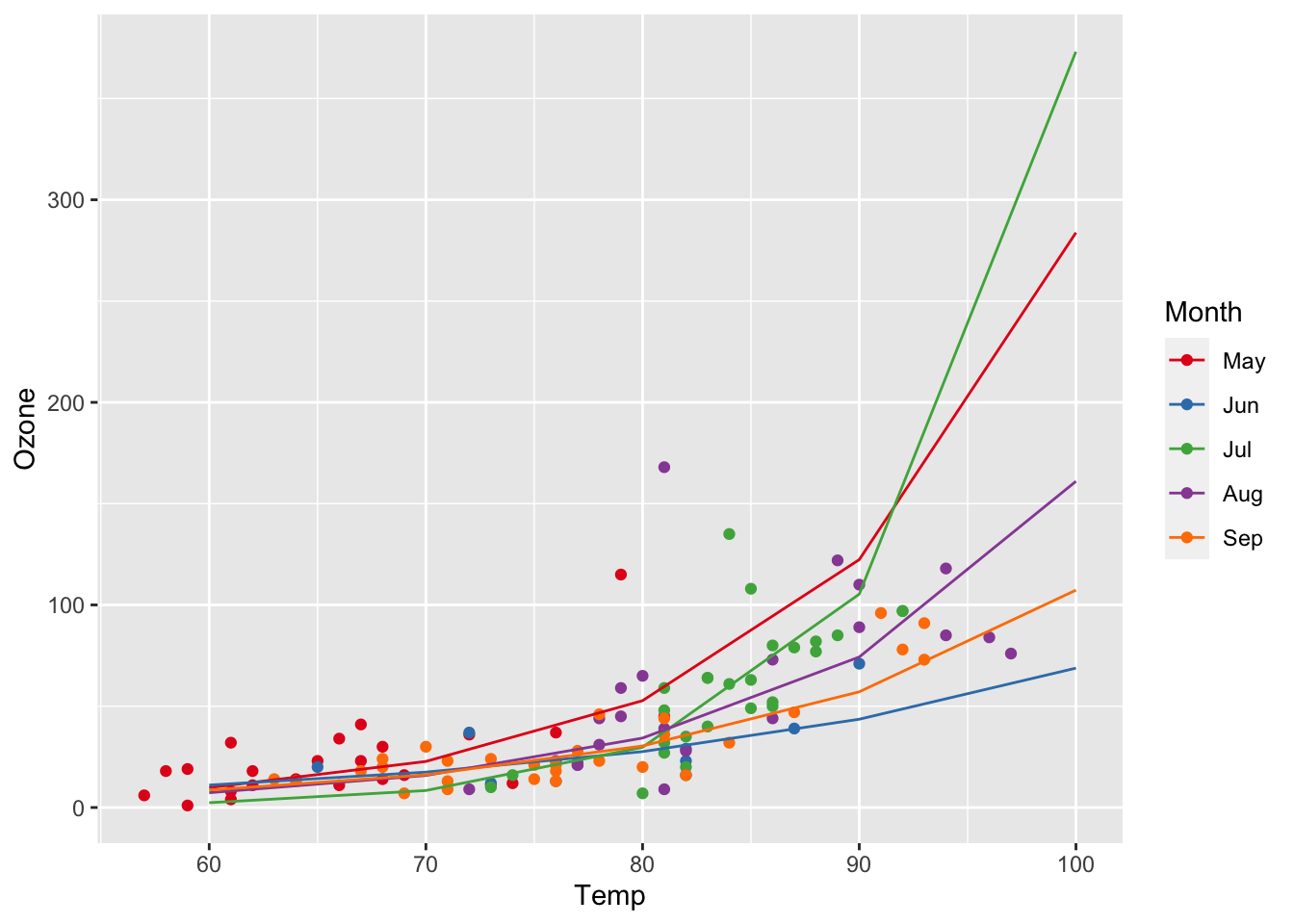
Resolving the simulation interval finer:
efct <- effect(term = "Temp:Month", mod = lm6,
xlevels = list(Temp = seq(57, 97, length.out = 100))) # Changing the x-levels
efct <- as.data.frame(efct)
efct$Month <- factor(efct$Month, levels = c("May", "Jun", "Jul", "Aug", "Sep"))
ggplot(airq, aes(x = Temp, y = Ozone, col = Month)) +
geom_point() +
scale_color_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00")) +
geom_line(data = efct, aes(y = exp(fit)))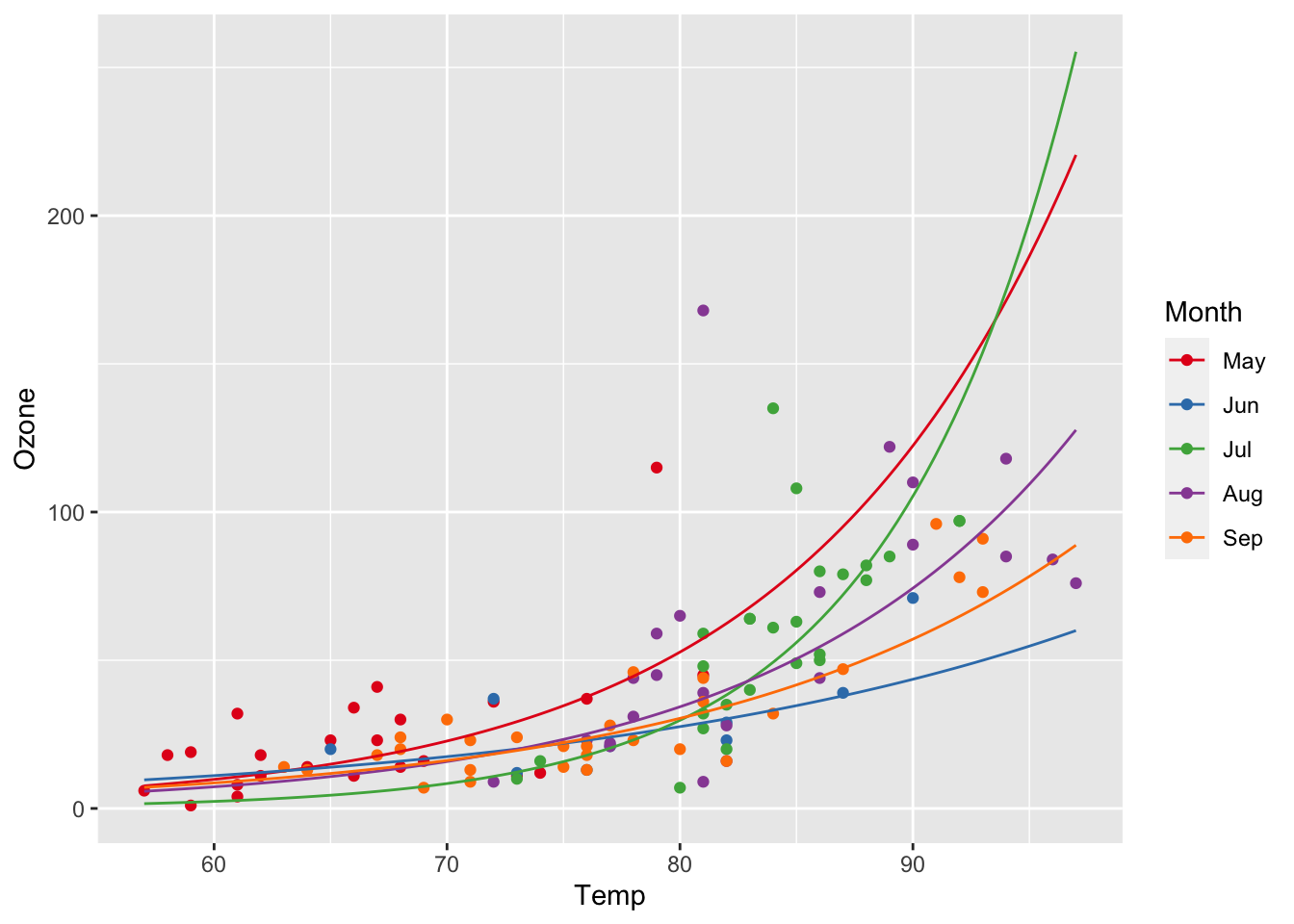
And finally with confidence intervals:
ggplot(airq) +
geom_point(aes(x = Temp, y = Ozone, col = Month)) +
geom_ribbon(data = efct, aes(x = Temp, ymin = exp(lower), ymax = exp(upper), fill = Month), alpha = 0.2) +
geom_line(data = efct, aes(x = Temp, y = exp(fit), col = Month)) +
scale_color_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00")) +
scale_fill_manual(values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00"))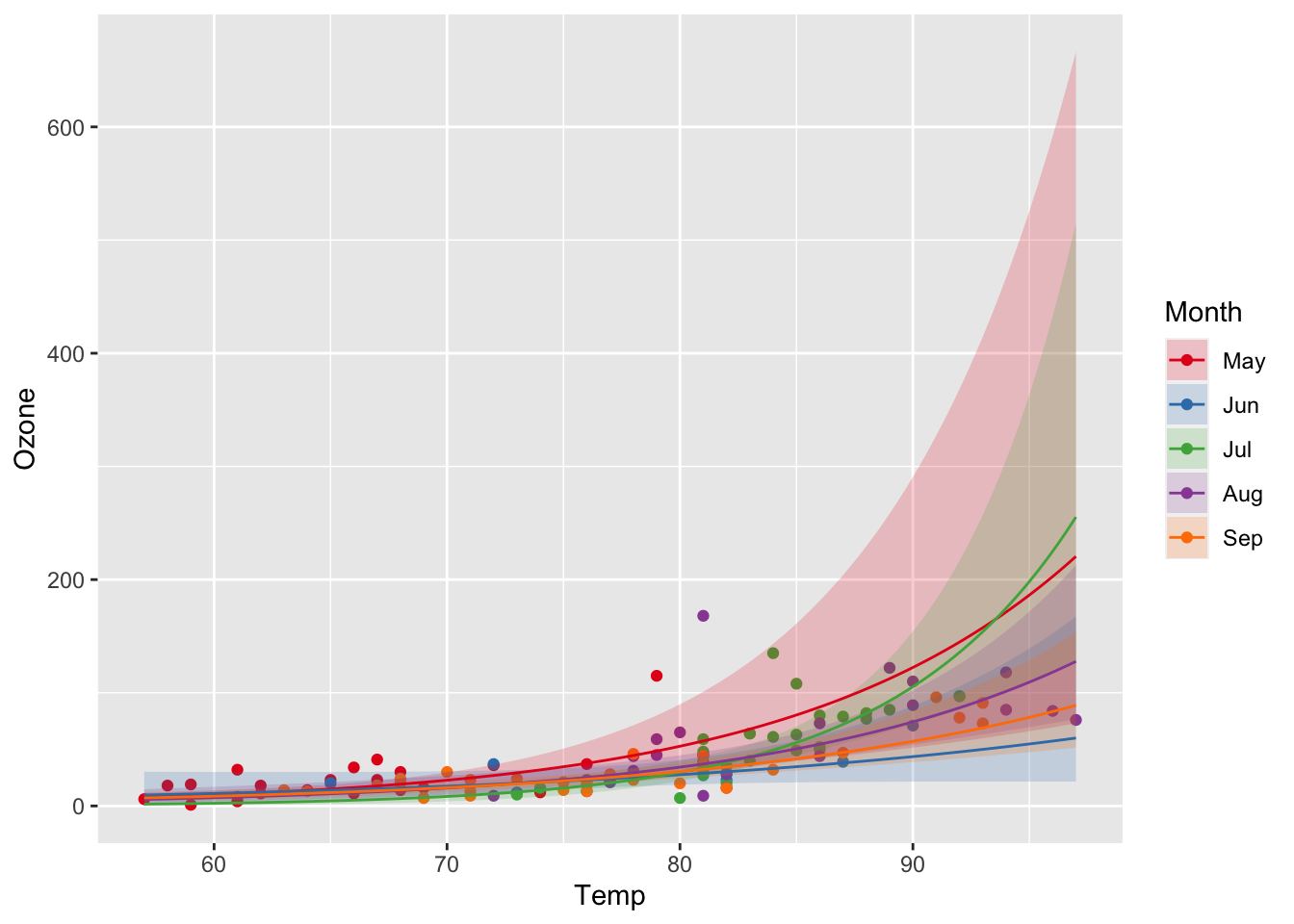
We can also specify an interaction between two continuous variables:
lm7 <- lm(logOzone ~ Temp * Wind, data = airq)summary(lm7)##
## Call:
## lm(formula = logOzone ~ Temp * Wind, data = airq)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.31151 -0.28199 0.05219 0.34752 1.12276
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.380182 1.306868 -1.821 0.0714 .
## Temp 0.081641 0.015953 5.118 1.37e-06 ***
## Wind 0.135132 0.115136 1.174 0.2431
## Temp:Wind -0.002509 0.001465 -1.713 0.0896 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.5475 on 107 degrees of freedom
## Multiple R-squared: 0.6111, Adjusted R-squared: 0.6002
## F-statistic: 56.04 on 3 and 107 DF, p-value: < 2.2e-16(Intercept): logOzone concentration at 0 windspeed and 0 degrees temperature. If the variables windspeed and temperature are standardised (z-transformed) then the intercept can be interpreted as logOzone concentration at average windspeed and average temperature, which is sometimes more intuitive.
Temp: Increasing temperature by 1 degree F at a windspeed of 0 increases logOzone concentration by 0.08.
Wind: Increasing windspeed by 1 mph is associated with an increase in logOzone concentration of 0.14, when temperature is 0.
Temp:Wind: The effect of temperature on logOzone concentration decreases by -0.003 for every unit increase in windspeed. For example, at zero windspeed, the effect (slope) of temperature on logOzone concentration is 0.082. At a windpeed of 1 mph, the effect decreases to 0.082 - 0.003 = 0.079. Here, the interpretation of the interaction term as windspeed modulating the temperature effect is physically more realistic, while mathematically the interpretation also works the other way round.
Let’s start with the formula showing the additive terms for temperature (β1Temp) and wind (β2Wind) and the interaction term between temperature and wind (β3TempWind):
logOzone=β0+β1Temp+β2Wind+β3Temp∗Wind+ϵ
We want to calculate the regression line between temperature and ozone for different levels of windspeed. We can rearrange the equation as follows:
y=(β0+β2Wind)+(β1+β3Wind)∗Temp+ϵ
Now we can say that the intercept is (β0+β2Wind) and the slope for temperature is (β1+β3Wind).
We can graph the interaction using the effects package:
efct <- effect("Temp:Wind", mod = lm7,
xlevels = list(Temp = seq(57, 97, length.out = 100)))
efct <- as.data.frame(efct)
ggplot(airq) +
geom_point(aes(x = Temp, y = Ozone, col = Wind)) +
geom_line(data = efct, aes(x = Temp, y = exp(fit), col = Wind, group = Wind))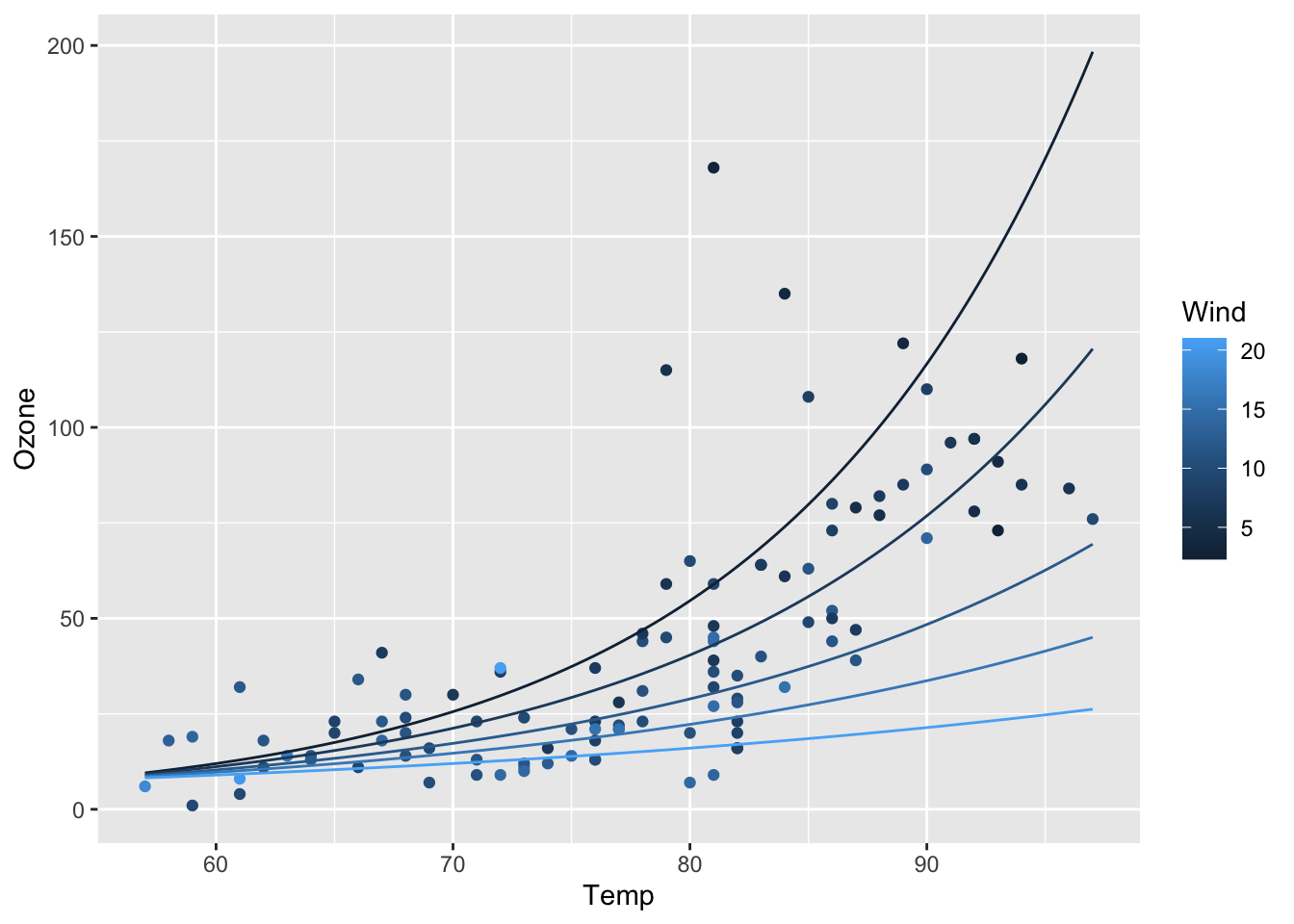
We can also change the values of the modulating variable within the
xlevels argument:
efct <- effect("Temp:Wind", mod = lm7,
xlevels = list(Temp = seq(57, 97, length.out = 100),
Wind = c(0, 10, 20, 30)))
efct <- as.data.frame(efct)
ggplot(airq) +
geom_point(aes(x = Temp, y = Ozone, col = Wind)) +
geom_line(data = efct, aes(x = Temp, y = exp(fit), col = Wind, group = Wind))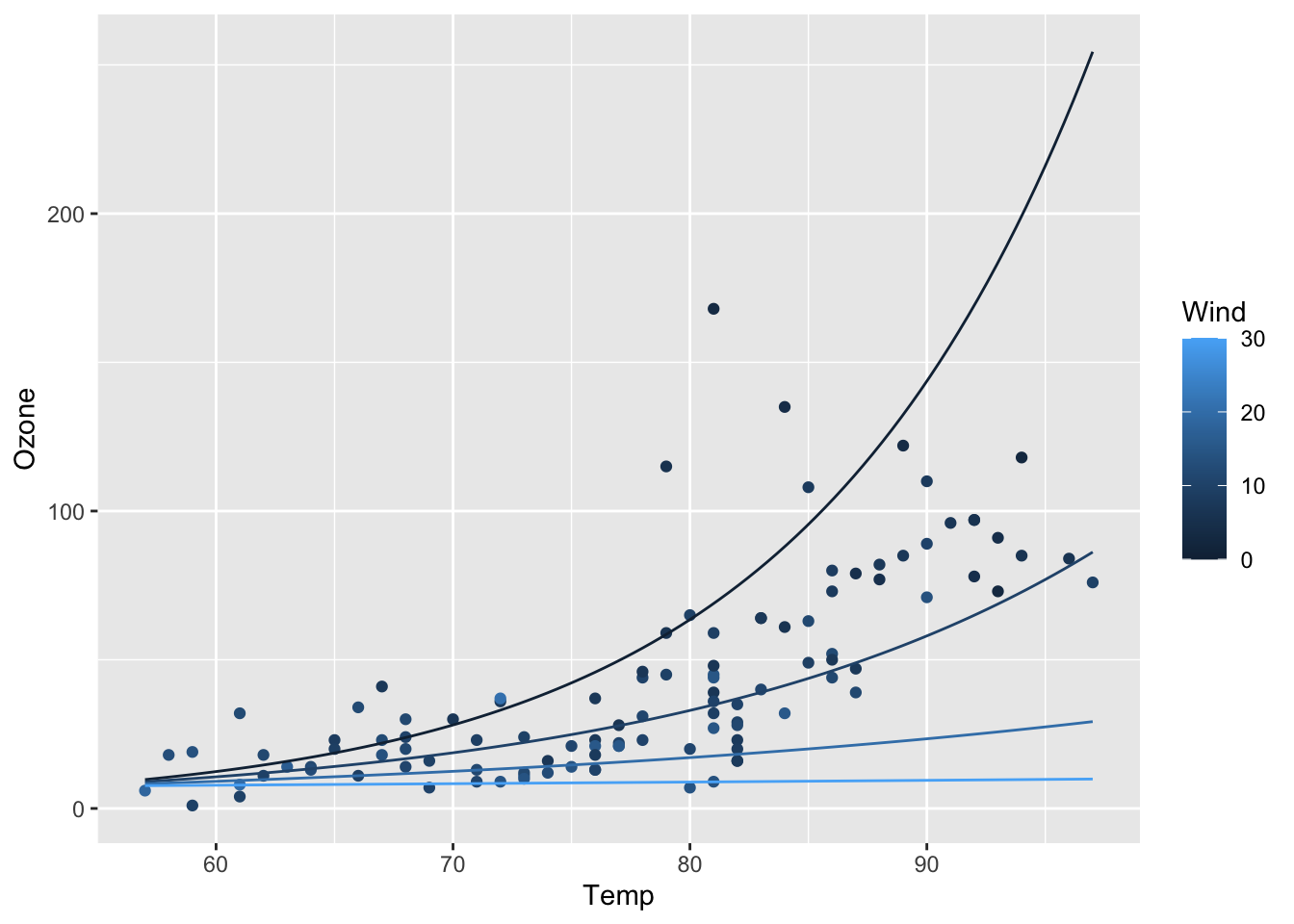
For interpretation, it is often much better to scale the predictors to unit scale (via z-transformation):
airq[, 3:5] <- scale(airq[, 3:5]) # z-transforming Solar, Wind, Temp
lm8 <- lm(logOzone ~ Temp * Wind, data = airq)
summary(lm8)##
## Call:
## lm(formula = logOzone ~ Temp * Wind, data = airq)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.31151 -0.28199 0.05219 0.34752 1.12276
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3.37401 0.05744 58.739 < 2e-16 ***
## Temp 0.54037 0.06017 8.980 1.03e-14 ***
## Wind -0.21364 0.06017 -3.551 0.000572 ***
## Temp:Wind -0.08507 0.04967 -1.713 0.089649 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.5475 on 107 degrees of freedom
## Multiple R-squared: 0.6111, Adjusted R-squared: 0.6002
## F-statistic: 56.04 on 3 and 107 DF, p-value: < 2.2e-16Given the interpretation above, the coefficient for Temp now indicates the increase in logOzone with an increase in temperature by 1 standard deviation (9.5 F) at mean windspeed (9.9 mph).
Again plotting the regression model:
efct <- effect("Temp:Wind", mod = lm8,
xlevels = list(Temp = seq(-2, 2, length.out = 100)))
efct <- as.data.frame(efct)
ggplot(airq) +
geom_point(aes(x = Temp, y = Ozone, col = Wind)) +
geom_line(data = efct, aes(x = Temp, y = exp(fit), col = Wind, group = Wind))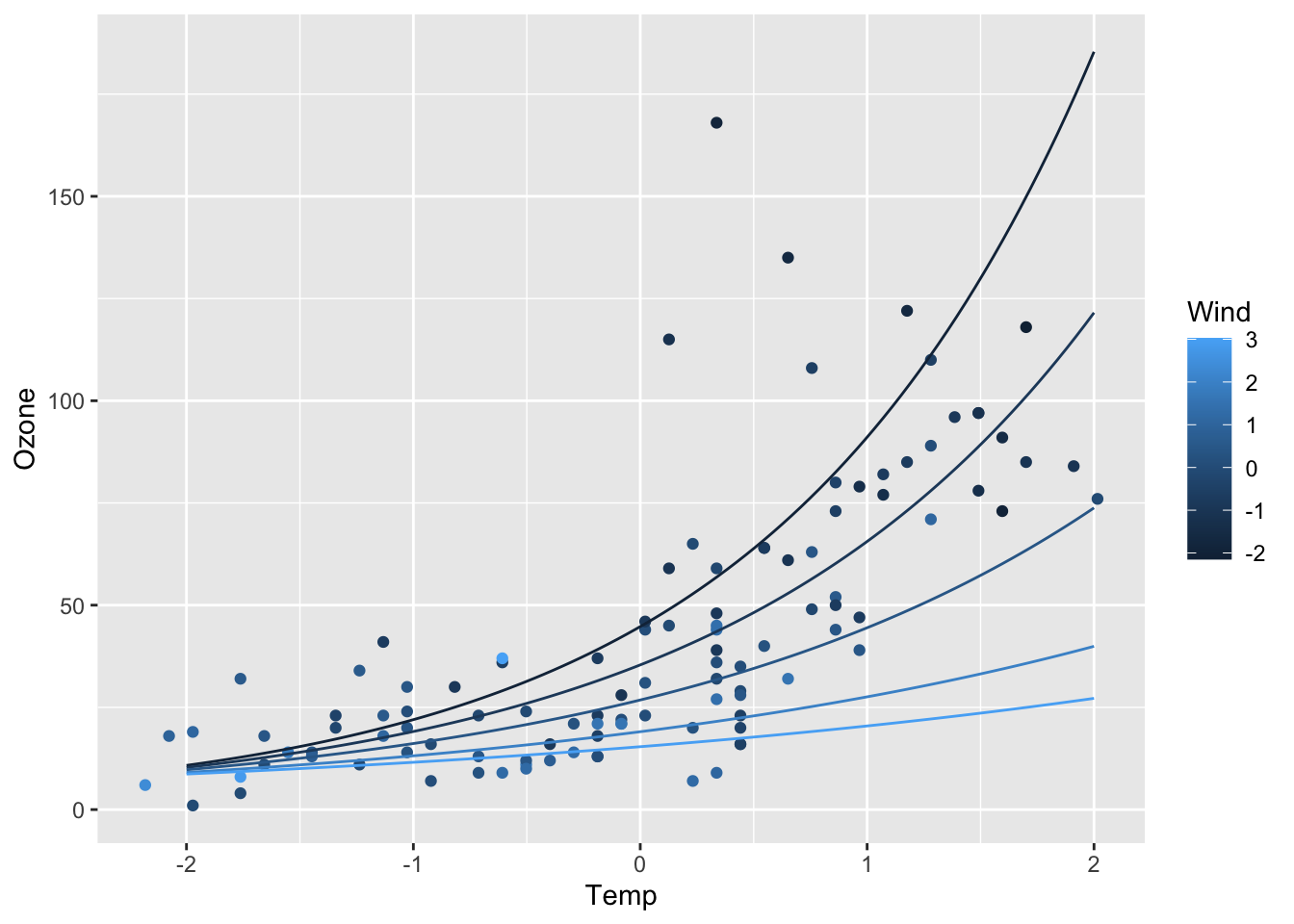
The Akaike Information Criterion (AIC) balances the goodness of fit with a penalty for model complexity. AIC is defined such that the smaller the value of AIC the better the model. Compare models based on the Akaike Information Criterion:
AIC(lm1, lm2, lm3)## df AIC
## lm1 5 170.8251
## lm2 6 168.5865
## lm3 7 168.0743What is the advantage of AIC over R2?
c(summary(lm1)$r.squared, summary(lm2)$r.squared, summary(lm3)$r.squared)## [1] 0.6644239 0.6769963 0.6842246c(summary(lm1)$adj.r.squared, summary(lm2)$adj.r.squared, summary(lm3)$adj.r.squared)## [1] 0.6550153 0.6648075 0.6691876R has several functions for performing model searches, e.g. based on the Akaike Information Criterion. Such functions make the process of model selection efficient, particularly when there are lots of predictor variables. In the seminar, we looked at manual backward and forward model selection:
Backward elimination:
Forward elimination:
One advantage of doing model reductions by hand is that you can impose some logical structure on the process, based on your understanding of the data and the underlying processes.
Here, we want to show you an alternative approach to backward/forward
model selection: exhaustive model comparison. Exhaustive model
comparison makes use of the power of modern computers to compare all
possible model combinations. For doing so, we use the function
dredge() in the package MuMIn:
options(na.action = "na.fail") # Prevent fitting models when data set changes due to NAs
fit_full <- lm(logOzone ~ (Temp * Wind + Solar) * Month, data = airq)
fit_all <- MuMIn::dredge(global.model = fit_full, rank = AIC)The result is a list of competing models:
head(fit_all, n = 5)## Global model call: lm(formula = logOzone ~ (Temp * Wind + Solar) * Month, data = airq)
## ---
## Model selection table
## (Int) Mnt Slr Tmp Wnd Mnt:Tmp Mnt:Wnd Tmp:Wnd Mnt:Tmp:Wnd df logLik AIC delta
## 143 3.367 0.2365 0.4681 -0.2197 -0.09930 6 -77.967 167.9 0.00
## 496 3.717 + 0.2307 0.7637 -0.5348 + + -0.36290 + 22 -63.100 170.2 2.27
## 15 3.416 0.2293 0.4686 -0.2190 5 -80.413 170.8 2.89
## 80 3.482 + 0.1954 0.4944 -0.1406 + 13 -73.074 172.1 4.21
## 208 3.469 + 0.2002 0.4919 -0.2188 + -0.06214 14 -72.498 173.0 5.06
## weight
## 143 0.569
## 496 0.183
## 15 0.134
## 80 0.069
## 208 0.045
## Models ranked by AIC(x)The first model in this list is the best according to AIC and then remaining models are ranked from next best to worst by difference in AIC, ΔAIC, with respect to the best model. Some of these lower-rank models are quite similar to each other in terms of AIC, i.e. model 2 and model 3 in this list, and model 4 and model 5.
We can extract the best model according to AIC like this:
model_selection <- get.models(fit_all, delta < 2) # Returns a list of models, in our case only one!
model <- model_selection[[1]]
model##
## Call:
## lm(formula = logOzone ~ Solar + Temp + Wind + Temp:Wind + 1,
## data = airq)
##
## Coefficients:
## (Intercept) Solar Temp Wind Temp:Wind
## 3.3670 0.2365 0.4681 -0.2197 -0.0993Copyright © 2024 Humboldt-Universität zu Berlin. Department of Geography.