- Data
- Fit PCA
- Scores
- Rotation coefficients
- Eigenvalues
- Correlation analysis
- Biplot
prcompvsprincomp
Principal Component Analysis
Humboldt-Universität zu Berlin | Geography Department
Data
WorldClim is a database of high spatial resolution global weather and climate data that can be used for mapping and spatial modeling. The bioclim dataset dervies from worldclim, i.e., from monthly temperature and rainfall values to generate more biologically meaningful variables. Bioclim are often used in species distribution modeling and related ecological modeling techniques. The original bioclim dataset contains 19 layers. For this demonstration, we’ll use a random sample collected over a geographic subset and only a subset of the variables:
BIO1 = Annual Mean Temperature
BIO5 = Max Temperature of Warmest Month
BIO6 = Min Temperature of Coldest Month
BIO12 = Annual Precipitation
clim <- read.csv("Data/bioclim_subset_sample.csv")
head(clim)## Tmean Tmax Tmin Precip
## 1 8.483958 21.75500 -7.096000 705.5000
## 2 6.497375 20.52900 -9.607000 749.0000
## 3 10.452375 24.45300 -5.697000 660.5000
## 4 8.237074 22.02089 -8.008889 637.1111
## 5 8.276444 21.41000 -6.908667 621.8333
## 6 11.461416 25.75200 -4.883000 684.2500A look at the scatter plot matrix shows that Tmean and
Tmax are strongly correlated with each other and that both
variables are also correlated to some degree with Tmin. If
we were to use these four variables in a regression model, we would face
the issue of multicollinearity. Statistical inference for regression
models hat contain intercorrelations among two or more independent
variables is less reliable as multicolinearity leads to wider confidence
intervals. In this case, it will likely be difficult to tell whether an
effect is due to Tmean or Tmax. One way to reduce multicollinearity is
to eliminate variables based on correlation coefficients or scientific
knowledge. Often this is an effective approach if there is prior
knowledge regarding the variables, but it can be cumbersome for larger
sets of predictors. Another approach is the principal component
analysis, which reduces the dimensionality (less predictors are needed)
and it yields uncorrelated predictors.
pairs(clim)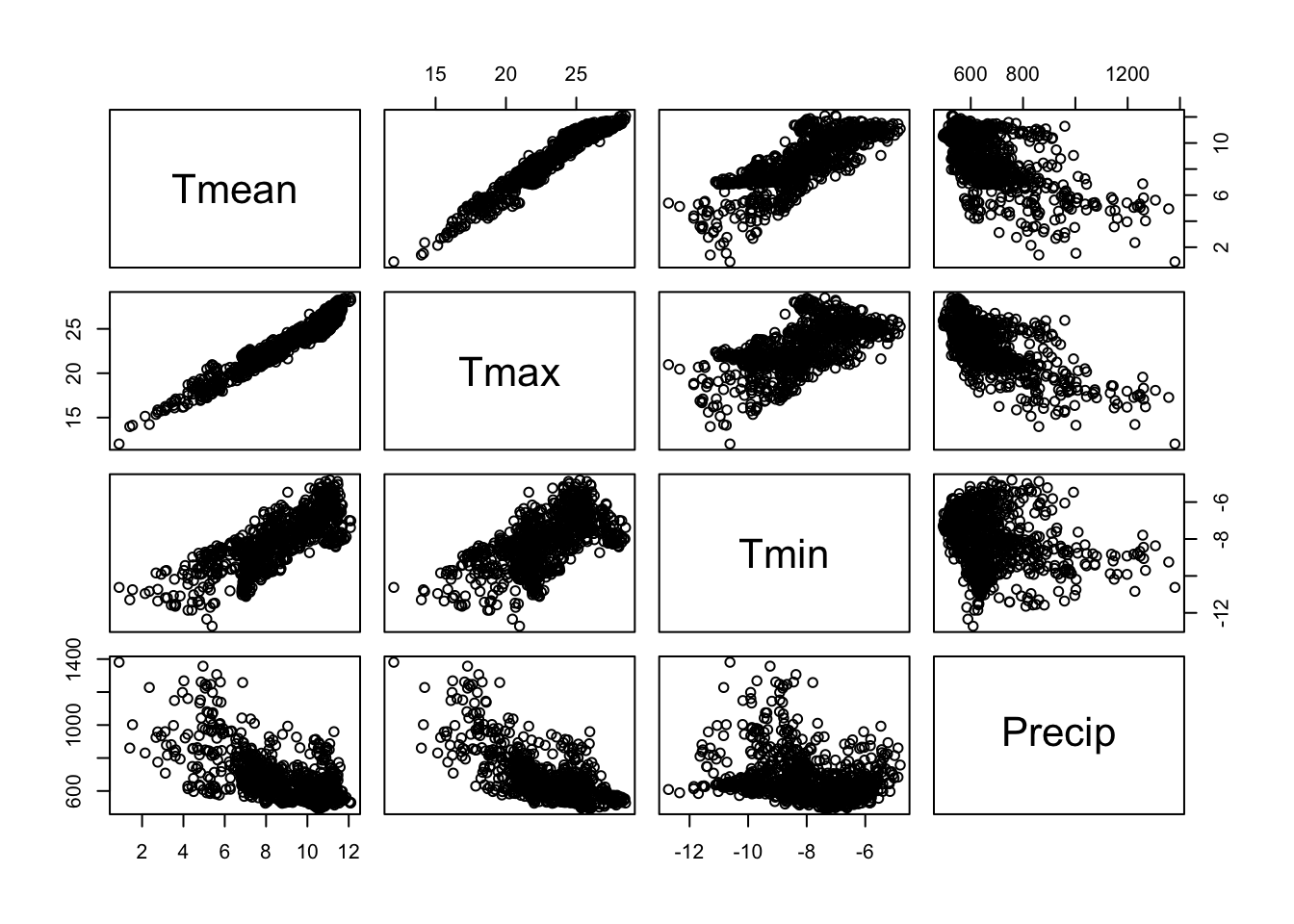
Fit PCA
There are two functions in R for performing a PCA:
princomp() and prcomp(). While the former
function calculates the eigenvalue on the correlation/co-variance matrix
using the function eigen(), the latter function uses
singular value decomposition, which numerically more stable. We here are
going to use prcomp().
pca <- prcomp(clim, scale. = TRUE)You should set the scale. argument to TRUE,
to scale the data matrix to unit variances. The default is
scale. = FALSE. You may also use scale() to
standardize the original data matrix and then enter the standardized
data matrix to prcomp.
Scores
The prcomp object stores the transformation and the
transformed values, which are called scores. You can access the
transformed dataset as follows:
pca_scores <- data.frame(pca$x)
head(pca_scores)## PC1 PC2 PC3 PC4
## 1 -0.10437038 -0.6623921 0.5218369 -0.06333806
## 2 -1.81424024 0.2309886 -0.1090850 0.02493079
## 3 1.57278761 -1.0321770 0.4015154 0.02441915
## 4 -0.19815948 0.1256635 0.3803971 -0.05908909
## 5 0.06685627 -0.2714205 1.0023861 -0.04971012
## 6 2.30800977 -1.5264628 0.2270009 0.07219499You see the transformed variables (now called PC1, PC2, PC3, PC4) are uncorrelated.
pairs(pca_scores)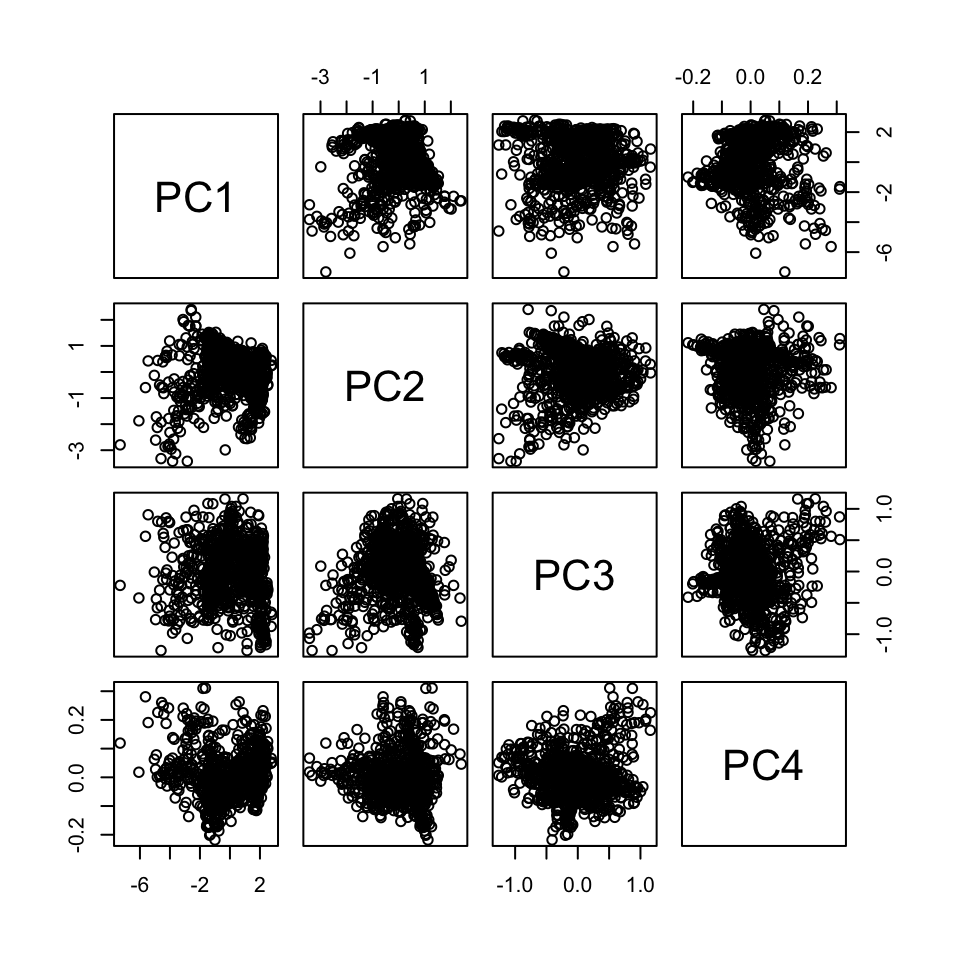
You can apply the transformation (rotation coefficients) to a dataset
using the predict function. In the example below, the
transformation is applied to the same dataset that was used to fit the
transformation. Hence, the results are the same as the scores stored in
the prcomp object (pca$x).
predicted_pca_scores <- predict(pca, clim)
head(predicted_pca_scores)## PC1 PC2 PC3 PC4
## [1,] -0.10437038 -0.6623921 0.5218369 -0.06333806
## [2,] -1.81424024 0.2309886 -0.1090850 0.02493079
## [3,] 1.57278761 -1.0321770 0.4015154 0.02441915
## [4,] -0.19815948 0.1256635 0.3803971 -0.05908909
## [5,] 0.06685627 -0.2714205 1.0023861 -0.04971012
## [6,] 2.30800977 -1.5264628 0.2270009 0.07219499Rotation coefficients
The PCA transformation consists of rotation coefficients
(eigenvectors). You can access the rotation coefficients stored in a
prcomp object as follows:
pca$rotation## PC1 PC2 PC3 PC4
## Tmean 0.5671218 -0.1334564 -0.2842325 -0.76142902
## Tmax 0.5561077 0.0899659 -0.5600202 0.60747649
## Tmin 0.4561343 -0.6196440 0.5977105 0.22522181
## Precip -0.4013209 -0.7682036 -0.4983286 0.02175513Recall, the coefficients describe how each PC is a linear combination of the input variables. An interesting analogy is a cooking recipe. The cooking recipe below defines the fractions of the climate variables that are needed (and added up) to “cook up” the first principle component.
PC1=0.567∗Tmean+0.556∗Tmax+0.456∗Tmin+−0.401∗Precip Let’s do this calculation for the
first observation in our bioclim dataset. Note, that below I scale the
variables to unit variance and mean zero (z-score). When
running the principal component analysis above, I set the scale argument
of the prcomp function to TRUE, which has the
same effect.
# first observation
clim_obs <- head(scale(clim),1)
clim_obs## Tmean Tmax Tmin Precip
## [1,] -0.07088585 -0.4483495 0.6604827 0.2893138# rotation coefficients of the first PC
pca1_coeff <- pca$rotation[,1]
pca1_coeff## Tmean Tmax Tmin Precip
## 0.5671218 0.5561077 0.4561343 -0.4013209Multiplying the observation vector and the coefficient vector is a vectorized operation. That is, the first element of the first vector is multiplied with the first element of the second vector, the second element of the first vector is multiplied with the second element of the second vector, etc.
sum(clim_obs * pca1_coeff)## [1] -0.1043704We just calculated the score of the first principle component (PC1)
for a single observation. The PC1 is a transformation (the cooking
recipe) and the score is the value you get when applying the
transformation. Let’s compare our score with the one obtained with
prcomp.
head(pca$x, 1)## PC1 PC2 PC3 PC4
## [1,] -0.1043704 -0.6623921 0.5218369 -0.06333806So, the coefficients reflect the contribution of each variable to each principal component. For example, the absolute magnitude of the PC1 coefficients for Tmean, Tmax, Tmin, and Precip is relatively similar, which indicates that all four variables contribute (roughly) similarly to PC1.
The sign of the coefficients tells us something about the direction, i.e., PC1 increases with increasing Tmean, Tmax, and Tmin, whereas PC1 decreases with increasing Precip. Note, however, the direction of the eigenvector is arbitrary. The equation below flips the signs of the coefficients (direction of the eigenvector). The inverse: −1∗PC1 is equally valid:
PCiˆ=−PCi
So, do not get attached to the direction of the PC1. But what the signs tell us is: Tmean, Tmax, and Tmin have an opposite sign to Precip. We can also say: Tmean, Tmax, and Tmin form a contrast to Precip (while one has a positive effect the other has a negative effect or vice versa). Now, to interpret what that means requires scientific knowledge of the underlying ecological, climatological, biological, or socio-ecological gradients you might be observing here, e.g. elevation gradients, continentality, etc.
Eigenvalues
To explore how the total variance of our four climate variables is
now re-partitioned in the derived principal components, we need to take
a look a the eigenvalues. Recall, the eigenvalues are the variances
explained by the principal components (one for each component). The
square roots of the eigenvalues are stored in the prcomp
object:
pca$sdev## [1] 1.73331374 0.88945906 0.44527838 0.07882397From this, we can easily calculate the variance explained by each component (the component variance divided by the total variance).
var_exp <- data.frame(pc = 1:4,
var_exp = pca$sdev^2 / sum(pca$sdev^2))
# add the variances cumulatively
var_exp$var_exp_cumsum <- cumsum(var_exp$var_exp)
var_exp## pc var_exp var_exp_cumsum
## 1 1 0.751094132 0.7510941
## 2 2 0.197784356 0.9488785
## 3 3 0.049568208 0.9984467
## 4 4 0.001553304 1.0000000The so called scree plot plots the component variances versus the component number. The idea is to look for an “elbow” which corresponds to the point after which the eigenvalues decrease more slowly.
par(mfrow=c(1,2))
plot(var_exp$pc, var_exp$var_exp, type="b", col=4,
ylab="Fraction of explained variance", xlab="Principle Component")
plot(var_exp$pc, var_exp$var_exp_cumsum, type="b", col=6,
ylab="Cummulative fraction variance", xlab="Principle Component")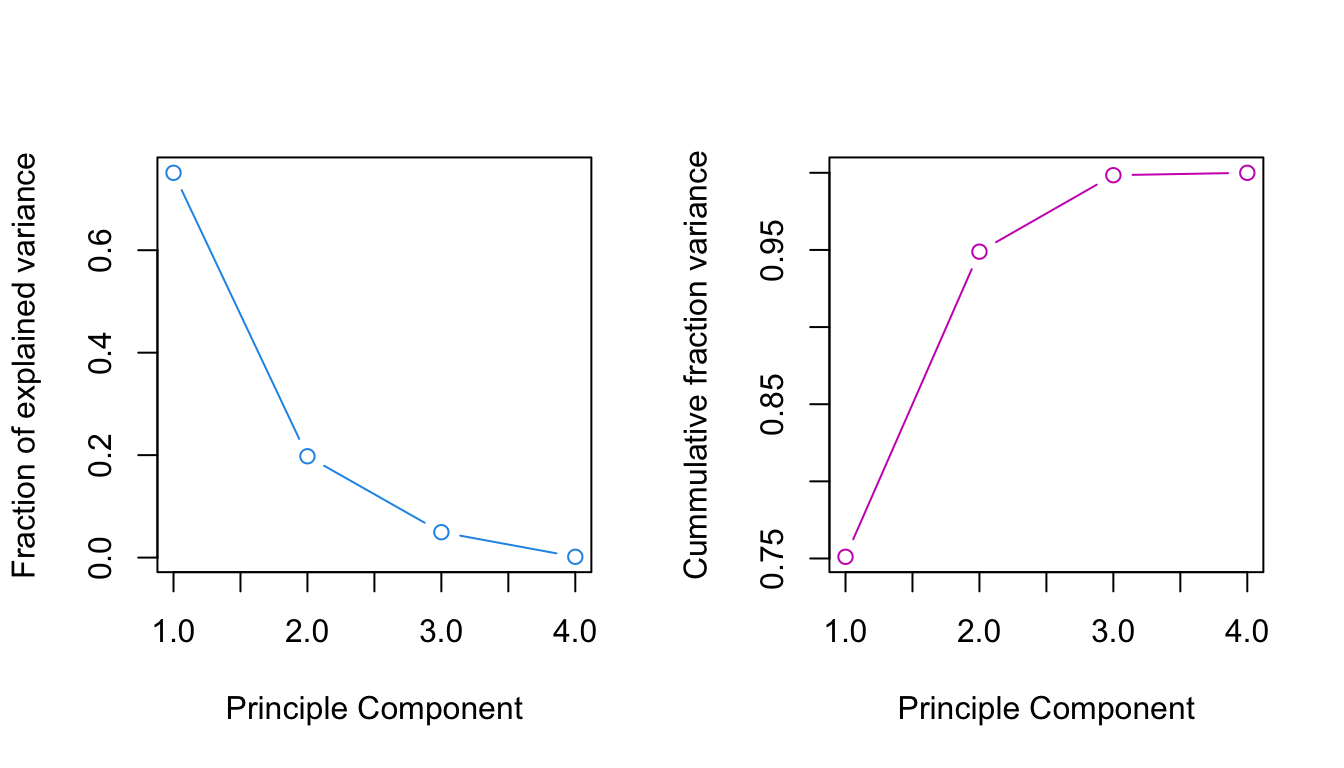
You are probably not surprised there is an easier way:
summary(pca)## Importance of components:
## PC1 PC2 PC3 PC4
## Standard deviation 1.7333 0.8895 0.44528 0.07882
## Proportion of Variance 0.7511 0.1978 0.04957 0.00155
## Cumulative Proportion 0.7511 0.9489 0.99845 1.00000The eigenvalues tell us: 75.1% of the variance is explained by PC1 alone, 19.8% by PC2, 5% by PC3, and only 0.16% by PC4. The cumulative proportion makes clear that 99.8% of the variance is captured by the first three components. Hence, we may discard PC4 without loosing much information.
Correlation analysis
Another way to interpret the meaning of the principal components is to explore the correlation between the PC scores and the original variables. Explore the scatter plot below and look at how PC1 is correlated with the orginal variables. Compare this to your observation of the rotation coefficients. You can do the same for PC2, PC3, and PC4.
df <- cbind(clim, pca_scores)
pairs(df[,1:7])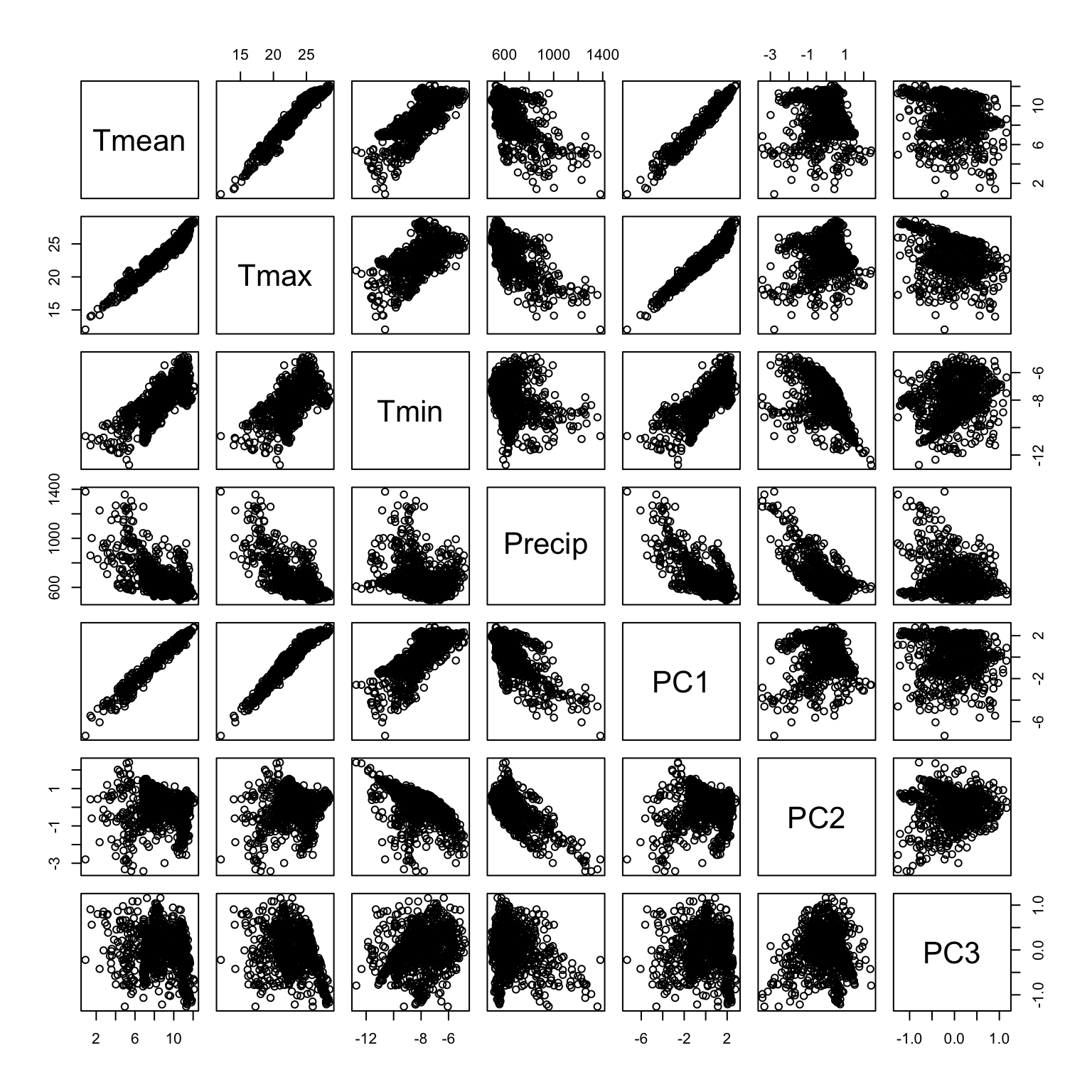
Biplot
The biplot shows the bioclim variables (in form of the rotation coefficients / loadings) and the observations (the PCA scores of our 10k samples) in one graphic. The variables are visualized as arrows from the plot origin, reflecting their direction and magnitude. Note, there are different versions of biplots out there also in different packages (e.g. PCAtools, ggbiplot).
biplot(pca)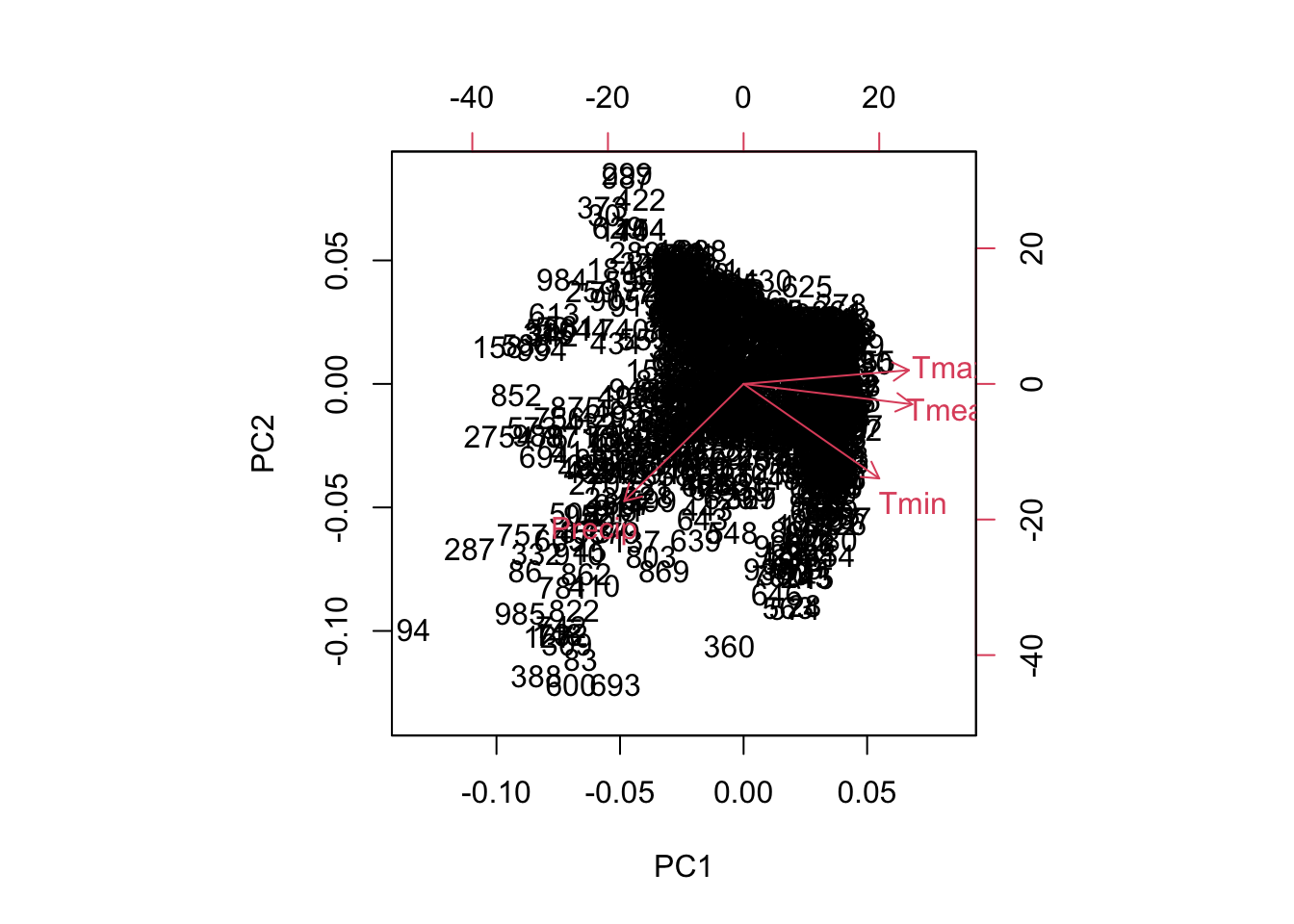
biplot(pca, choices=c(1,3))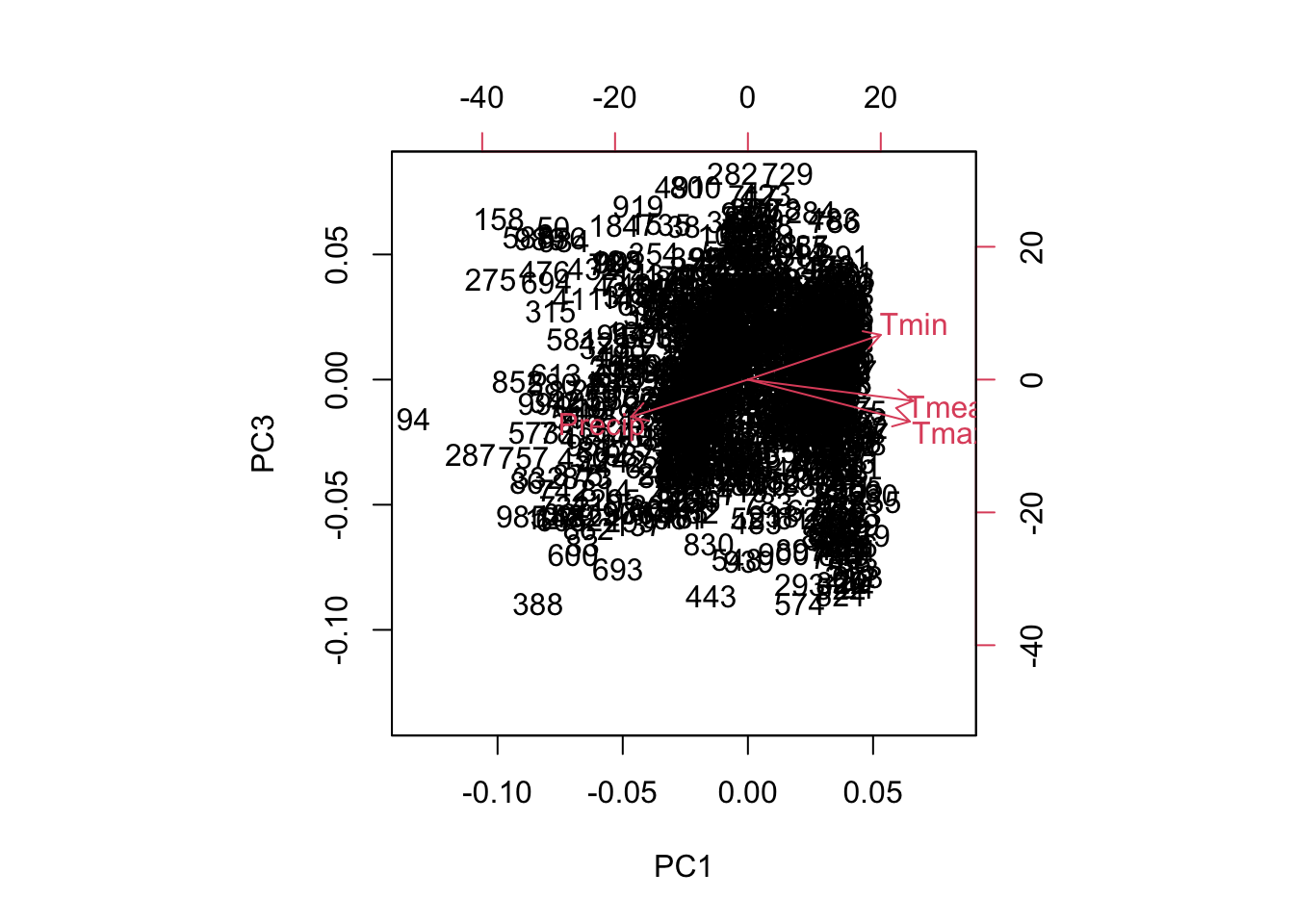
biplot(pca, choices=c(1,3), xlabs=rep('.', nrow(pca$x)))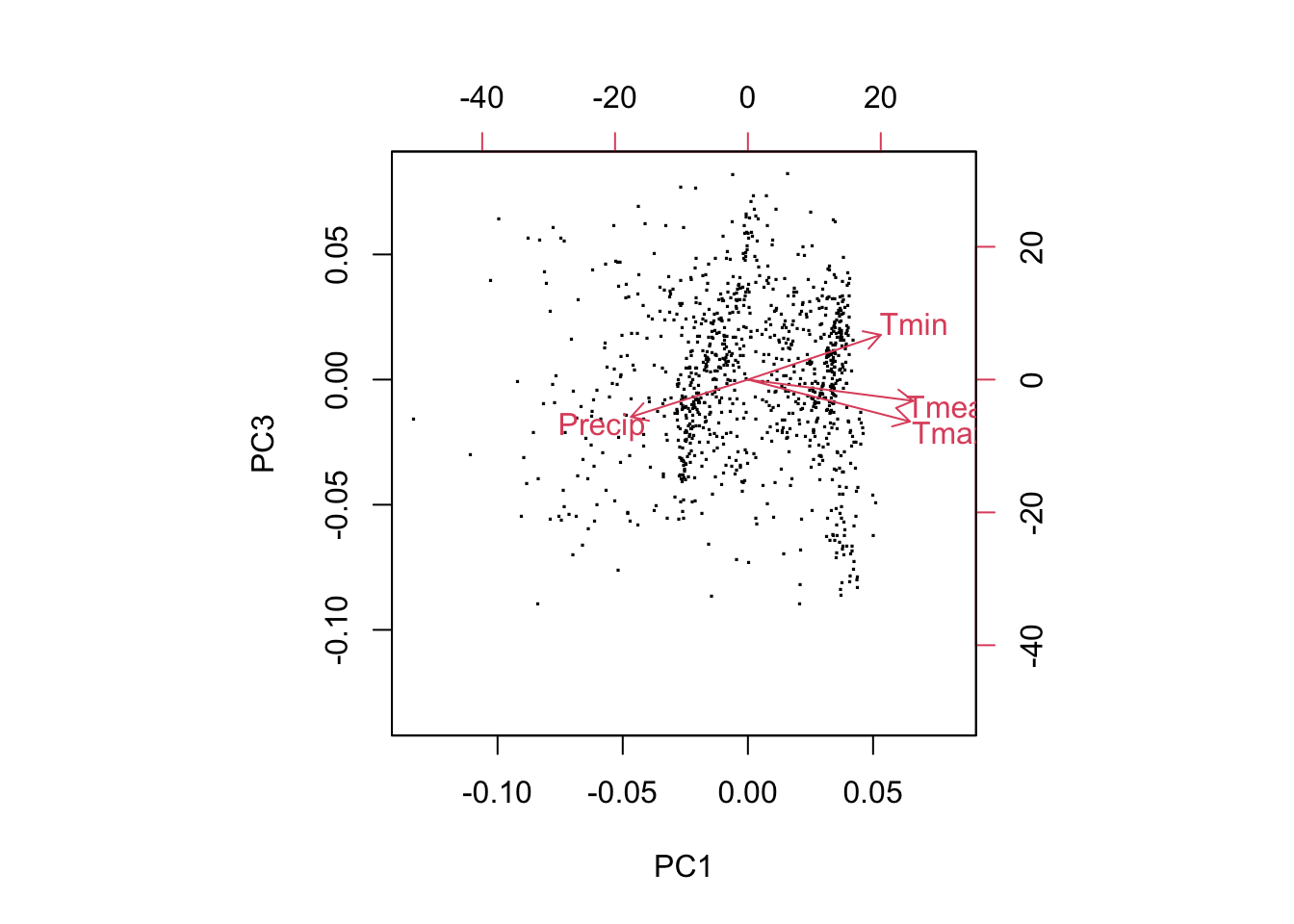
prcomp vs princomp
| Description | prcomp() | princomp() |
|---|---|---|
| Decomposition method | svd() | eigen() |
| Standard deviations of the principal components | sdev | sdev |
| Matrix of variable loadings (columns are eigenvectors) | rotation | loadings |
| Variable means (means that were substracted) | center | center |
| Variable standard deviations (the scalings applied to each variable ) | scale | scale |
| Coordinates of observations on the principal components | x | scores |
Copyright © 2024 Humboldt-Universität zu Berlin. Department of Geography.