- Learning goals
- Change detection principles
- Bi-temporal change detection
- Overall Workflow
- Exercise
- Land Cover Mapping
- Change Detection
- Aggregating classes
- Validating the change map
- Summarizing key results
- Assignment: Prepare Project Work
10. Change Detection
Learning goals
- Understand the principles of change detection
- Conduct bi-temporal change detection of forest cover in the Brazilian Amazon
Change detection principles
Remote sensing provides great opportunities to observe and monitor changes on the land surface. Using satellite based images, changes over long periods of time as well as over large areas can be revealed. This section aims at providing insights into the most basic form of change detection - the bi-temporal change detection.
Bi-temporal change detection
To illustrate the procedure for bi-temporal change detection, we consider a hypothetical example of deforestation mapping. We would like to assess changes in forest cover, thereby revealing potential forest loss, gain, and stability.
We are given two images for the same study site, one from 1992 and one from 2011.
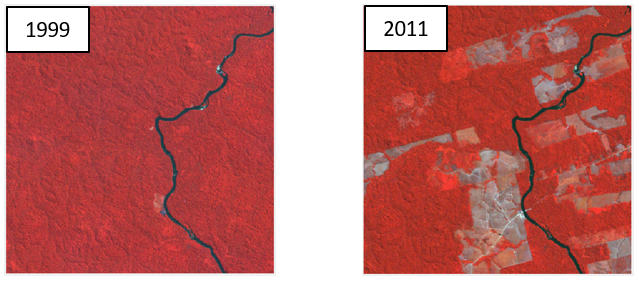
For each image we want to produce a binary map showing forest / non-forest. To do so, we collect two sets of training data points: one for both classes in each image. Next, we use supervised classification techniques (e.g. Random Forest) to create one map per image. In this case we train one classification model for each image.
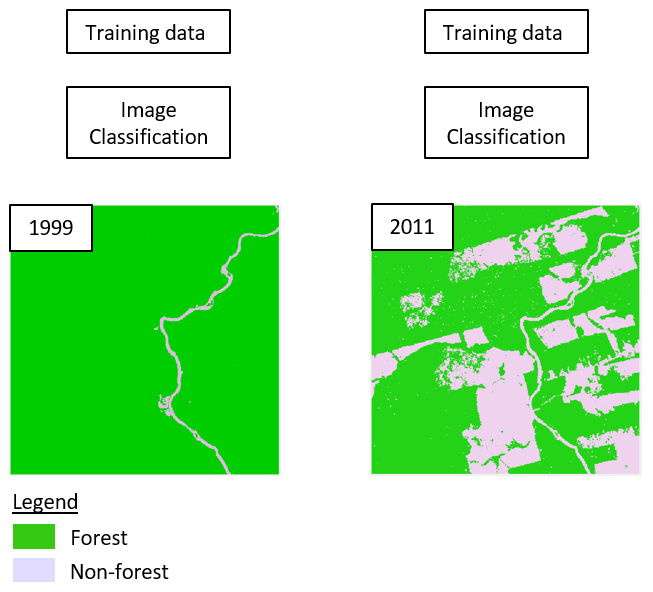
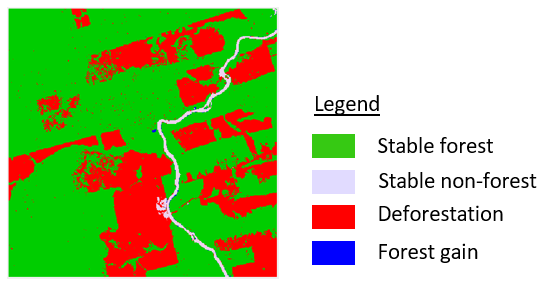
As a next step, we want to derive change information from the two maps. We can therefore combine the maps in order to assess class trajectories for each pixel. Remember, the classes in the map correspond to numeric values, e.g. 1 (forest) and 2 (non-forest). Since we observe bi-temporal change of a binary (2 classes) map, we can have classestimesteps=22=4 different sequences and thus change classes.
This map allows us to assess if change has occurred or not. In the binary case we can observe two types of stable sites
- Stable forests: pixels with class label forest in 1992 and forest in 2011
- Stable non-forests: pixels with class label non-forest in 1992 and non-forest in 2011
Also, we can observe two change classes
- Deforestation: pixels with class label forest in 1992 and non-forest in 2011
- Forest gain: pixels with class label non-forest in 1992 and forest in 2011
Or in tabular form:
| Class 1992 | Class 2011 | Change class description | Change class ID |
|---|---|---|---|
| 1 | 1 | Stable Forest | 11 |
| 2 | 2 | Stable Non-Forest | 22 |
| 1 | 2 | Deforestation | 12 |
| 2 | 1 | Forest Gain | 21 |
To create a third map that contains these four classes, we can use a simple mathematical trick: The most straightforward is to multiply the pixel values of the first image with 10 and then add the pixel values of the second image. As an example:
- Stable forest (forest to forest): 1 * 10 + 1 = 11
- Deforestation (forest to non-forest): 1 * 10 + 2 = 12
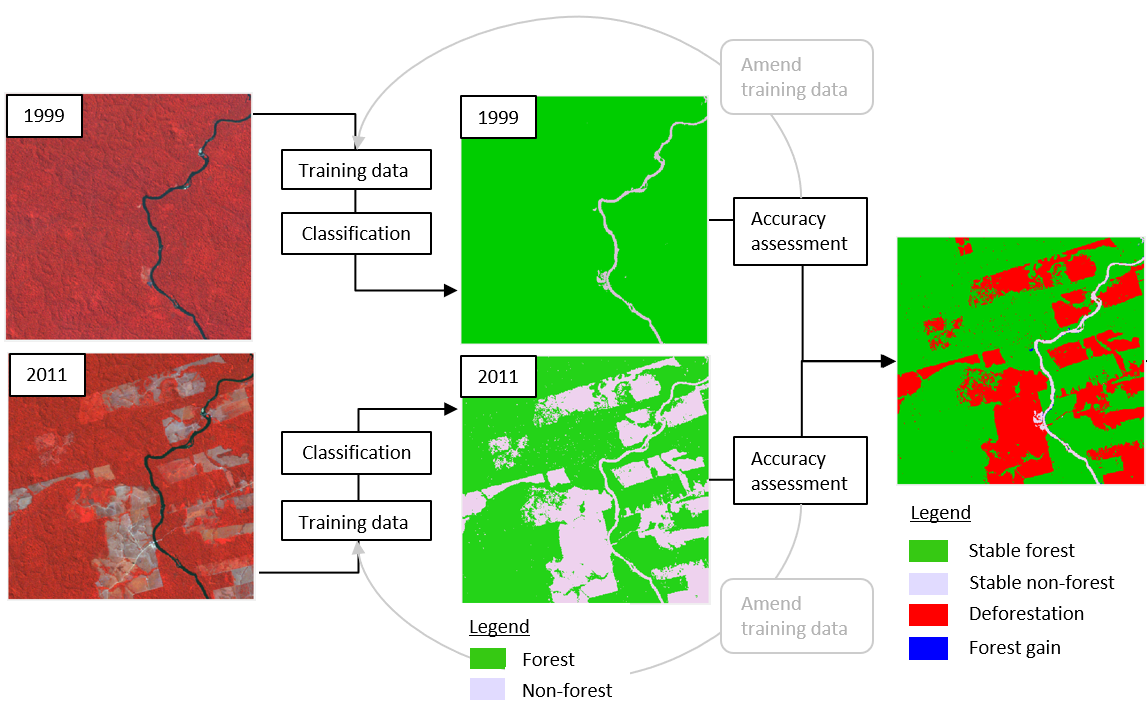
Overall Workflow
Let’s put this whole process of creating the maps and assessing change into a single workflow. It can be divided in six steps and summarized graphically as follows:
Exercise
Download the session materials from our shared repository.
The exercise for today deals with implementing this entire process chain, with a focus on the change detection (i.e. the classification process is already pre-coded and you only have to copy and comprehend the code).
The following datasets are available to you:
- Training data
training_brazil_forest.gpkg: “Forest” (classID:1) and “Non-Forest” (classID:2) reference points for both timesteps - Image features
LT5_227065_19920808_subset.tifandLT5_227065_20110718_subset.tif: Two Landsat-5 images from 1992 and 2011
Land Cover Mapping
We will repeat the classification workflow from Session 8 and Session 9:
- Data preparation: Extract the feature values of the
Landsat-5 image at the training point locations into a
data.frame - Model building: Supply the feature-label data.frame to the
randomForest-function - Validation (internal model evaluation using Out-Of-Bag (OOB) error)
- Prediction: Classify the Landsat-5 image using the image data,
model, and the
terra::predictfunction
The only difference being here that we have to build two models and also predict land cover twice. Here is the code to achieve this:
library(randomForest)
# input data
img_1992 <- rast()
img_2011 <- rast()
train_forest <- vect()
# rename bandnames
bandnames <- c("BLU", "GRN", "RED", "NIR", "SW1", "SW2")
names(img_1992) <- bandnames
names(img_2011) <- bandnames
# 1992
df_1992 <- data.frame(
terra::extract(x=img_1992, y=train_forest["LC_1992"], bind=TRUE)
)
df_1992$LC_1992 <- as.factor(df_1992$LC_1992)
df_1992 <- na.omit(df_1992)
rf_1992 <- randomForest(LC_1992~., df_1992, ntree=500)
print(rf_1992)
# 2011
df_2011 <- data.frame(
terra::extract(x=img_2011, y=train_forest["LC_2011"], bind=TRUE)
)
df_2011$LC_2011 <- as.factor(df_2011$LC_2011)
df_2011 <- na.omit(df_2011)
rf_2011 <- randomForest(LC_2011~., df_2011, ntree=500)
print(rf_2011)
# Prediction
cl_1992 <- terra::predict(img_1992, rf_1992)
cl_2011 <- terra::predict(img_2011, rf_2011)randomForest model outputs
(print(rf_1992) and print(rf_2011)). What is
the overall accuracy according to the OOB error? What are the User’s and
Producer’s accuracy for forest and non-forest in each model?
Change Detection
With both classifications in place we can transition to mapping the change classes, using the above mentioned trick to create the change map.
Use image arithmetic in R to create a change map showing all possible (no) change classes following the procedure outlined above (it is just one line of code). Plot the result to verify it worked and write the resulting raster to disc.
change_map <-
plot(change_map)
writeRaster()Visualize the change map in QGIS and rename the change classes in your QGIS legend.
Aggregating classes
If we are - for instance - only interested in the deforestation mapping and would like to simplify the values of the corresponding raster back into a binary map, we can simply use indexing to create a new map. In this specific case we would like to aggregate non-relevant classes to class “others” using indexing: * 12 –> 1 = Deforestation * 11, 21, 22 –> 2 = Others (Stable Non-Forest, Stable Forest, Forest Gain)
One way to achieve this is to do the following:
change_map[change_map == 12] <- 1 # Deforestation becomes 1
change_map[change_map == 11 | change_map == 21 | change_map == 22] <- 2 # Others becomes 2Validating the change map
Accuracy assessment is an crucial step in any remote sensing-based
classification exercise, given that classification maps always contain
mis-classified pixels and, thus, classification errors. When we are
explicitly mapping change classes and want to report this change we need
an independent sound accuracy assessment. This should follow the
guidlines from last session and requires sampling validation data
explicitly for the change classes. We provide you with a validation
sample
(validation/brazil_deforestation_validation_bin_change.shp)
shapefile, but for time reasons we will skip this step today.
Summarizing key results
Calculate absolute (e.g. hectares/km2) and relative (% of study area) estimates that describe the change process (e.g. area deforested in hectares, or how much percent of the forest hast been deforested).
Absolute values: - Calculate the area in hectare of a single Landsat pixel using the spatial resolution accessed from the metadata of the raster - Identify only the pixels that were deforested and multiply by the area of a single pixel
Relative values: - Divide the deforested area estimate from above by the total area of the raster
pixel_area_hectares <-
total_deforested_hectares <-
total_area_hectares <-
percent_deforested <-
print(paste("Total deforested area: ", total_deforested_hectares, "ha"))
print(paste("Percent of study area deforested: ", percent_deforested, "%"))Assignment: Prepare Project Work
Portfolio
Next session we will start into the Project Phase.
Preparation for Portfolio:
- Read and comprehend the MAP project (note down questions regarding general points)
- Read through the three paper’s under consideration of the questions posed in the portfolio
Copyright © 2023 Humboldt-Universität zu Berlin. Department of Geography.